Incidence of BCR-ABL transcript variants in patients with chronic myeloid leukemia: Their correlation with presenting features, risk scores and response to treatment with imatinib mesylate
CC BY-NC-ND 4.0 · Indian J Med Paediatr Oncol 2014; 35(01): 26-30
DOI: DOI: 10.4103/0971-5851.133707
Abstract
Context: The exact role of the different transcript variants of BCR-ABL in the pathogenesis of chronic myeloid leukemia (CML) and their impact on prognosis is yet to be definitely enumerated. Aims: In this study, we have tried to correlate the presenting features, risk scores and treatment response with the BCR-ABL variants detected in our patients. Settings and Design: A cross-sectional unicentric hospital-based study on 80 patients diagnosed to have CML by bone marrow cytogenetics and confirmed by reverse transcriptase-polymerase chain reaction (RT-PCR). Materials and Methods: RT-PCR for BCR-ABL was performed on consecutive patients with CML attending the CML clinic from January 2010 to December 2010.The medical charts of these patients were analyzed after a follow-up of 18 months in a retrospective manner. Statistical Analysis: Box plot and histogram was used to see the distribution of variables. t-test was performed to enumerate the difference between risk scores in two populations of patients carrying two different BCR-ABL transcript variants. Results: Nearly 56.25% of patients had b3a2 (e14a2) while 41.25% of patients showed b2a2 (e13a2) transcripts. The rest 2.5% (two patients) expressed the rare e19b2 variant. Patients with b2a2 presented with higher Sokal, Hasford and European Treatment and Outcomes Study score than their b3a2 counterpart. Different parameters such as the platelet count, leukocyte count, hemoglobin and splenomegaly showed a minor difference between the groups. More patients in the b2a2 group achieved complete hematologic response at 3 months, but it was not significant. Conclusions: Patients with b2a2 variant CML tend to present with higher risk score, but do not behave in a vastly different manner than their b3a2 counterparts.
Keywords
BCR-ABL transcript variants - chronic myeloid leukemia - chronic myeloid leukemia risk scores - imatinib mesylatePublication History
Article published online:
19 July 2021
© 2014. Indian Society of Medical and Paediatric Oncology. This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial-License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used for commercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/.)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
Abstract
Context:
The exact role of the different transcript variants of BCR-ABL in the pathogenesis of chronic myeloid leukemia (CML) and their impact on prognosis is yet to be definitely enumerated.
Aims:
In this study, we have tried to correlate the presenting features, risk scores and treatment response with the BCR-ABL variants detected in our patients.
Settings and Design:
A cross-sectional unicentric hospital-based study on 80 patients diagnosed to have CML by bone marrow cytogenetics and confirmed by reverse transcriptase-polymerase chain reaction (RT-PCR).
Materials and Methods:
RT-PCR for BCR-ABL was performed on consecutive patients with CML attending the CML clinic from January 2010 to December 2010. The medical charts of these patients were analyzed after a follow-up of 18 months in a retrospective manner.
Statistical Analysis:
Box plot and histogram was used to see the distribution of variables. t-test was performed to enumerate the difference between risk scores in two populations of patients carrying two different BCR-ABL transcript variants.
Results:
Nearly 56.25% of patients had b3a2 (e14a2) while 41.25% of patients showed b2a2 (e13a2) transcripts. The rest 2.5% (two patients) expressed the rare e19b2 variant. Patients with b2a2 presented with higher Sokal, Hasford and European Treatment and Outcomes Study score than their b3a2 counterpart. Different parameters such as the platelet count, leukocyte count, hemoglobin and splenomegaly showed a minor difference between the groups. More patients in the b2a2 group achieved complete hematologic response at 3 months, but it was not significant.
Conclusions:
Patients with b2a2 variant CML tend to present with higher risk score, but do not behave in a vastly different manner than their b3a2 counterparts.
INTRODUCTION
Philadelphia chromosome i.e., the chromosome originating from the reciprocal translocation between long arms of chromosome 9 and chromosome 22 t(9;22)(q34;q11), along with its product BCR-ABL oncogene is perhaps the most extensively studied oncoprotein. Since its discovery in 1960,[1] its different variants in different types of leukemia has been recorded and analyzed. In chronic myeloid leukemia (CML) BCR-ABL fusion oncogene translates chimeric protein p210, which exerts its oncogenic role by activating the tyrosine kinase, which does not block differentiation but leads to the development of malignancy by enhancing proliferation of myeloid cell line.[2] In CML, most of the cases of p210 constitutes principally of two variants: b3a2 or e14a2 and b2a2 or e13a2.[3,4] The b3a2 variant is produced by the fusion of exon 14 of BCR gene with exon 2 of ABL gene while the other variant, i.e., b2a2 is produced by fusion of exon 13 of BCR and exon 2 of ABL.[5] These transcripts variants are produced as the result of alternate splicing. Many studies had been performed to find interrelation between the particular variant of p210 BCR-ABL and the etiology, pathogenesis, prognosis of the disease. What our study constitutes is the endeavor to find the interrelation between these two principal transcript variants of p210 and its association with different aspects of the disease such as white blood cell (WBC) count, platelet count, organomegaly, prognostic markers of the CML (Sokal score,[6] Hasford/Euro score,[7] European Treatment and Outcomes Study (EUTOS) score[8]) along with the response to the modern day mainstay therapy of CML through Imatinib mesylate. reverse transcriptase polymerase chain reaction (RT-PCR) chosen as the method of choice to determine the transcript variant of BCR-ABL, is one of the most sensitive methods for this purpose.
MATERIALS AND METHODS
The study complied with the Declaration of Helsinki and was approved by the ethics committee of the participating institution.
Patients
A total of 80 consecutive patients who were diagnosed to be suffering from CML in our clinic over the period of January 2010 to December 2010 were selected for the study.
Sample collection
Blood samples were collected from 80 patients from the out-patient Department of Institute of Hematology and Transfusion Medicine, Medical College Kolkata. Samples were collected from peripheral circulation (3 ml for each patient), in a proper aseptic manner, into properly labeled ethylenediaminetetraacetic acid vacuum tubes. Processing of the collected blood, i.e., beginning of ribonucleic acid (RNA) isolation from the collected blood sample, was started immediately, thus evading any chance of messenger RNA degradation. Proper consent from each patient was taken at the time of sample collection.
RNA isolation
Total RNA was isolated from a blood sample by using the QIAGEN RNA isolation kit (Qiagen, Germany) by following the protocol provided by the manufacturer. RNA quantity was determined by spectrophotometry.
Complementary DNA (cDNA) synthesis
RNA was reverse transcribed into cDNA for using as template in PCR reaction. RT reaction was performed by using Transcriptor First Strand cDNA Synthesis Kit, Roche applied science, USA.
6 μL of RNA (~2.4 μg RNA) was added to 14 μL of mix containing 2 μL of 10x RT reaction buffer (40 mM Tris-HCL, 100 mM KCl, 20 mM DTT), 10 mM dNTP, 10 mM oligo-dT, 20 units of RNase inhibitor and 40 units of Reverse Transcriptase. Next the mix was incubated in 42°C for 45 min, 99°C for 3 min and finally held at 4°C for 20 min. Manufacturer provided RNA is used as a positive control.
PCR amplification
The primers of standardized PCR protocol of BIOMED1 (Europe against Cancer)[9] was used to amplify the cDNA (Except the shifted primer ABL-a3-E3’). The sequences of the primers are given as follows:
- BCR-b1-A 3086 (22): GAAGTGTTTCAGAAGC TTCTCC
- ABL-a3-B 458 (21): GTTTGGGCTTCACACCATTCC
- BCR-b2-C 3126 (21): CAGATGCTGACCAACT CGTGT
- ABL-a3-D 441 (23): TTCCCCATTGTGATTA TAGCCTA.
The cDNA product of the former reaction was amplified with PCR reaction in two rounds (nested PCR). 150 mM primers, 1 U/ml Taq Polymerase, 10 mM dNTP mix and ×10 PCR buffer with 25 mM MgCl2 was used in each rounds. Initial denaturation was performed at 95°C for 3 min followed by 35 cycles of denaturation at 94°C for 20 s; annealing at 55°C for 15 s; extension at 72°C for 15 s with a total 35 cycles using thermocycler. Final extension was performed at 72°C for 5 min.
After performing the PCR reaction, the products were checked through agarose gel electrophoresis in 2% of agarose gel medium.
RESULT
The b3a2 transcript variant was recognized as 417 base pair long product and b2a2 was recognized as 342 base pair long product when primers A and B were used [Figure 1] and as 360 and 285 bp products when primers C and D were used. When nested PCR were performed, C and D, being internal primers were able to produce 360 and 285 products. The baseline characteristics of patients are shown in Table 1.
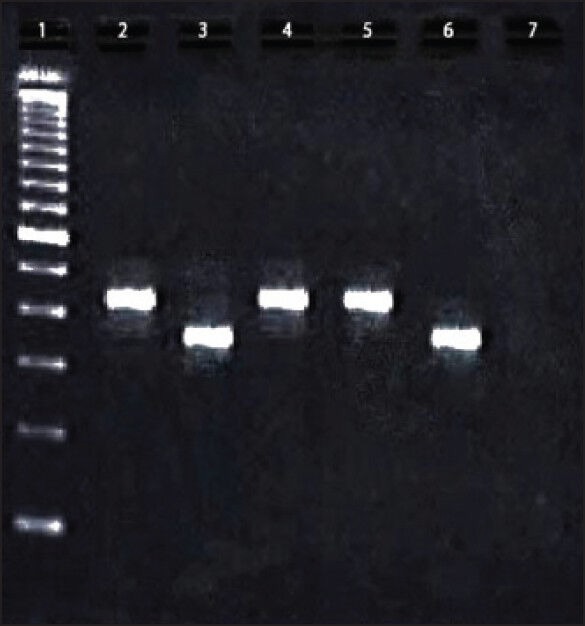
| Figure 1:Reverse transcriptase-polymerase chain reaction of BCR-ABL transcript variants in 2% agarose gel. Lane 1: 100bp ladder, Lane 2: Positive control of b3a2 (417bp); Lane 2: Positive control of b2a2 (342 bp); Lane 4 and 5: Patient sample showing b3a2 variant; Lane 6: Patient sample showing b2a2 variant; Lane 7: Negative control
Table 1
Baseline characteristics of patients
|
Amongst 80 patients (including 24 females) 56.25% of patients showed the presence of b3a2 and 41.25% of patients showed the presence of b2a2 transcript variant. The rest 2.5% (numerically, 2 cases) showed the expression of rare e19b2 variety, recognized as a 243 base pair long product in agarose gel electrophoresis. However, there was no case of b3a2 and b2a2 co-expression in any patient.
Bone marrow cytogenetics did not reveal any abnormality other than the presence of Philadelphia chromosome in any patient.
Qualitative nested PCR were performed on the patients at the end of the follow-up period as it was done at the beginning of the study. All the patients, including two patients who underwent complete molecular response, did show qualitative nested PCR value as positive at the end of the follow-up period (which is a common occurrence). Hence the first round PCR (at the time of diagnosis) and the second round PCR (at the end of follow-up) did not discriminate the patient population.
From the database of history of the patients, we had calculated the Sokal score, Hasford score and EUTOS score of all patients at the time of diagnosis and correlated them with the respective transcript variants of the patients is given in the Figure 2.
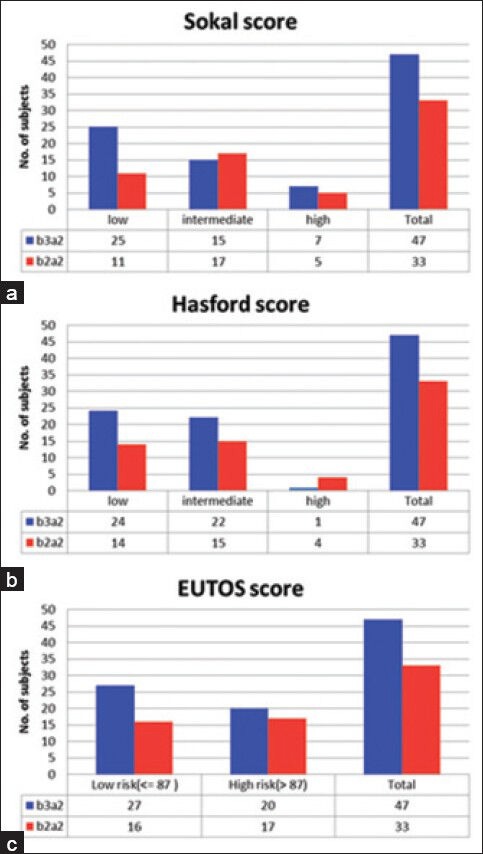
| Figure 2:B2a2 shows higher risk scores than b3a2 variety. Distribution of patients of both the transcript variants b3a2 and b2a2 are shown as in (a) Sokal score, (b) Hasford score and (c) European Treatment and Outcomes Study score described risk categories
From Figure 2, it is apparent that, according the Sokal, Hasford and EUTOS score, the patients with b2a2 transcript variants are presenting with higher scores than the patients having b3a2 variants. Hence we have performed t-test to test if the risk scores are significantly higher in patients with b2a2 variants and it was seen that the P < 0 xss=removed>P = 0.03 in case of Sokal score, 0.027 in EUTOS score and 0.24 in Hasford score.
We analyzed the correlation between different transcript variants and the progression of disease among the patients. We tabulated the transcript variants of the patients against their stage of the disease i.e., if they stayed in chronic phase (CP) all along or progressed to accelerated phase or blast crisis in Tables Tables22 and and33.
Table 2
Response to treatment
|
Table 3
Disease stage

|
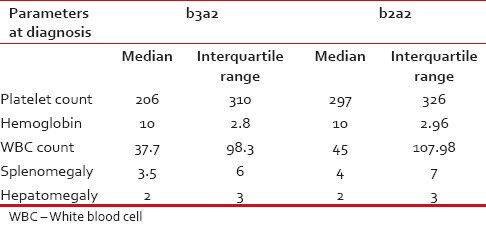
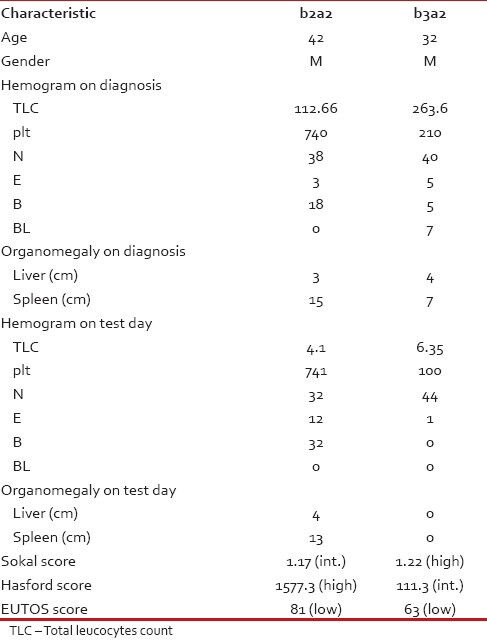
We have also tabulated the Platelet count, organomegaly and other hematological features against the transcript variants as there are earlier reports on the correlation between these two in the form of higher platelet count found in b3a2 variety.[10,11,12,13] However, we found higher platelet counts in patients with b2a2 variety of BCR-ABL than in b3a2. A summary of the two patients with e19a2 transcripts is depicted in Table 4.
Table 4
Clinical features of patients with transcript variants
|
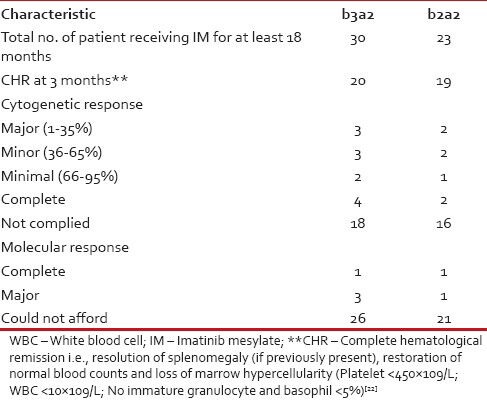
We also studied our patients’ response to the first generation tyrosine kinase inhibitor Imatinib mesylate and any possible relation with the transcript variants of the patients. For this purpose, we had only considered the data of the patients who presented to us in the CP of the disease (1st CP of CML-CP) and are currently being treated by Imatinib mesylate (at the standard dose of 400 mg/day) for at least a period of 6 months. In our population, we found 30 such cases of b3a2 and 23 such cases of b2a2. For assessment of the Imatinib response, we had to depend mostly on the hematological response as most of the patients could not afford the recommended bone marrow cytogenetics or quantitative RT-PCR (qRT-PCR). Cytogenetic response (partial, major or complete) and molecular response was assessed for BCR-ABL in a few cases where the patient could afford the test.
DISCUSSION
In our study, the frequency of b3a2 and b2a2 was found to be 56.25% and 41.25% respectively which is quite closer to the data derived in the similar studies performed in the Caucasian population.[11] No co-expression of these two variants was found in any of the patients. In other Asian countries, the distribution was also similar to our findings.[14,15] In all the aforementioned studies, the ratio was roughly calculated to be b3a2:b2a2 = 60:40. However, the role of BCR-ABL transcripts variants in the prognosis had always been a controversial issue. Although some authors reported no such role on the part of the transcript variants, others found data referring to the transcript variants to have prognostic values.[16,17,18] In our population, this is perhaps the first study on the transcript variants and their possible role on the presenting relative risks (measured by Sokal, Hasford and EUTOS score) and Imatinib response. Our study does not reveal a significant difference of treatment response in the two patient populations. The patients with b2a2 variants of our population seem to be presenting with higher relative risk (according to Sokal and EUTOS score). In the study by Martínez-Mancilla et al., a higher leukocyte count in b2a2 patients was reported.[19]
The presenting features of the patients of these two sub-populations are not starkly different either. Although some of the studies failed to show any correlation between platelet count and BCR-ABL transcript variants;[20] others recognized higher platelet count in patients carrying b3a2[10,11,12,13] However almost all the reported studies correlating these two, related higher platelet count in the b3a2 variety our study had revealed otherwise [Table 4]. The other presenting features as Total WBC count, Hemoglobin, hepatomegaly and splenomegaly are within moderate range of discrimination showing the b2a2 population presenting with relative organomegaly and slightly lower hemoglobin as well as lower WBC count.
Due to the issue of affordability of our patients, the world-wide recommended follow-up protocol through qRT-PCR to analyze transcript load of BCR-ABL every 3 months interval[21] could not be performed. Rather the common practice is to follow-up patients through checking for hematological remission. In addition in case of Imatinib failure, switching over to second generation tyrosine kinase inhibitor is impossible for the same reason of affordability. According to the criterion of achieving and maintenance of hematological remission, b2a2 patients seem to respond well to the Imatinib therapy. Still keeping the small sample size in mind, it is best to say that further investigation is required to conclude this aspect as well as to investigate the association between transcript variants and treatment response in terms of cytogenetic and molecular response.
Footnotes
Source of Support: Institutional Research fund and STS Indian Council of Medical Research.
Conflict of Interest: None declared.
References
- Nowell PC. The minute chromosome (Phl) in chronic granulocytic leukemia. Blut 1962;8:65-6.
- Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer 2005;5:172-83.
- Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science 1986;233:212-4.
- Lucas CM, Harris RJ, Giannoudis A, Davies A, Knight K, Watmough SJ, et al. Chronic myeloid leukemia patients with the e13a2 BCR-ABL fusion transcript have inferior responses to imatinib compared to patients with the e14a2 transcript. Haematologica 2009;94:1362-7.
- Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature 1985;315:550-4.
- Sokal JE, Cox EB, Baccarani M, Tura S, Gomez GA, Robertson JE, et al. Prognostic discrimination in "good-risk" chronic granulocytic leukemia. Blood 1984;63:789-99.
- Hasford J, Pfirrmann M, Hehlmann R, Allan NC, Baccarani M, Kluin-Nelemans JC, et al. A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon Alfa. Writing Committee for the Collaborative CML Prognostic Factors Project Group. J Natl Cancer Inst 1998;90:850-8.
- Hasford J, Baccarani M, Hoffmann V, Guilhot J, Saussele S, Rosti G, et al. Predicting complete cytogenetic response and subsequent progression-free survival in 2060 patients with CML on imatinib treatment: The EUTOS score. Blood 2011;118:686-92.
- van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: Investigation of minimal residual disease in acute leukemia. Leukemia 1999;13:1901-28.
- Perego RA, Costantini M, Cornacchini G, Gargantini L, Bianchi C, Pungolino E, et al. The possible influences of B2A2 and B3A2 BCR/ABL protein structure on thrombopoiesis in chronic myeloid leukaemia. Eur J Cancer 2000;36:1395-401.
- Verschraegen CF, Kantarjian HM, Hirsch-Ginsberg C, Lee MS, O′Brien S, Rios MB, et al. The breakpoint cluster region site in patients with Philadelphia chromosome-positive chronic myelogenous leukemia. Clinical, laboratory, and prognostic correlations. Cancer 1995;76:992-7.
- Inokuchi K, Inoue T, Tojo A, Futaki M, Miyake K, Yamada T, et al. A possible correlation between the type of bcr-abl hybrid messenger RNA and platelet count in Philadelphia-positive chronic myelogenous leukemia. Blood 1991;78:3125-7.
- Balatzenko G, Vundinti BR, Margarita G. Correlation between the type of bcr-abl transcripts and blood cell counts in chronic myeloid leukemia-A possible influence of mdr1 gene expression. Hematol Rep 2011;3:e3.
- Goh HG, Hwang JY, Kim SH, Lee YH, Kim YL, Kim DW. Comprehensive analysis of BCR-ABL transcript types in Korean CML patients using a newly developed multiplex RT-PCR. Transl Res 2006;148:249-56.
- Iqbal Z, Manzoor F, Iqbal M, Ali S, Sheikh N, Khan M. Frequency of BCR-ABL Fusion Oncogene Splice Variants Associated with Chronic Myeloid Leukemia (CML). J Cancer Ther 2011;2:176-80.
- de Lemos JA, de Oliveira CM, Scerni AC, Bentes AQ, Beltrão AC, Bentes IR, et al. Differential molecular response of the transcripts B2A2 and B3A2 to imatinib mesylate in chronic myeloid leukemia. Genet Mol Res 2005;4:803-11.
- Rosas-Cabral A, Martínez-Mancilla M, Ayala-Sánchez M, Vela-Ojeda J, Bahena-Reséndiz P, Vadillo-Buenfil M, et al. Analysis of Bcr-abl type transcript and its relationship with platelet count in Mexican patients with chronic myeloid leukemia. Gac Med Mex 2003;139:553-9.
- Udomsakdi-Auewarakul C, U-Pratya Y, Boonmoh S, Vatanavicharn S. Detection of molecular variants of BCR-ABL gene in bone marrow and blood of patients with chronic myeloid leukemia by reverse-transcriptase polymerase chain reaction (RT-PCR). J Med Assoc Thai 2000;83:928-35.
- Martínez-Mancilla M, Gutiérrez M, de la Rosa GZ, García-Carrancá A, Gariglio P, Miranda E. Younger age and shorter chronic phase in b2a2-positive chronic myeloid leukemia adults with high white blood cell count at diagnosis. Haematologica 2002;87:666-8.
- Shepherd P, Suffolk R, Halsey J, Allan N. Analysis of molecular breakpoint and m-RNA transcripts in a prospective randomized trial of interferon in chronic myeloid leukaemia: No correlation with clinical features, cytogenetic response, duration of chronic phase, or survival. Br J Haematol 1995;89:546-54.
- Baccarani M, Cortes J, Pane F, Niederwieser D, Saglio G, Apperley J, et al. Chronic myeloid leukemia: An update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol 2009;27:6041-51.
- Baccarani M, Saglio G, Goldman J, Hochhaus A, Simonsson B, Appelbaum F, et al. Evolving concepts in the management of chronic myeloid leukemia: Recommendations from an expert panel on behalf of the European LeukemiaNet. Blood 2006;108:1809-20.

| Figure 1:Reverse transcriptase-polymerase chain reaction of BCR-ABL transcript variants in 2% agarose gel. Lane 1: 100bp ladder, Lane 2: Positive control of b3a2 (417bp); Lane 2: Positive control of b2a2 (342 bp); Lane 4 and 5: Patient sample showing b3a2 variant; Lane 6: Patient sample showing b2a2 variant; Lane 7: Negative control

| Figure 2:B2a2 shows higher risk scores than b3a2 variety. Distribution of patients of both the transcript variants b3a2 and b2a2 are shown as in (a) Sokal score, (b) Hasford score and (c) European Treatment and Outcomes Study score described risk categories
References
- Nowell PC. The minute chromosome (Phl) in chronic granulocytic leukemia. Blut 1962;8:65-6.
- Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer 2005;5:172-83.
- Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science 1986;233:212-4.
- Lucas CM, Harris RJ, Giannoudis A, Davies A, Knight K, Watmough SJ, et al. Chronic myeloid leukemia patients with the e13a2 BCR-ABL fusion transcript have inferior responses to imatinib compared to patients with the e14a2 transcript. Haematologica 2009;94:1362-7.
- Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature 1985;315:550-4.
- Sokal JE, Cox EB, Baccarani M, Tura S, Gomez GA, Robertson JE, et al. Prognostic discrimination in "good-risk" chronic granulocytic leukemia. Blood 1984;63:789-99.
- Hasford J, Pfirrmann M, Hehlmann R, Allan NC, Baccarani M, Kluin-Nelemans JC, et al. A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon Alfa. Writing Committee for the Collaborative CML Prognostic Factors Project Group. J Natl Cancer Inst 1998;90:850-8.
- Hasford J, Baccarani M, Hoffmann V, Guilhot J, Saussele S, Rosti G, et al. Predicting complete cytogenetic response and subsequent progression-free survival in 2060 patients with CML on imatinib treatment: The EUTOS score. Blood 2011;118:686-92.
- van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: Investigation of minimal residual disease in acute leukemia. Leukemia 1999;13:1901-28.
- Perego RA, Costantini M, Cornacchini G, Gargantini L, Bianchi C, Pungolino E, et al. The possible influences of B2A2 and B3A2 BCR/ABL protein structure on thrombopoiesis in chronic myeloid leukaemia. Eur J Cancer 2000;36:1395-401.
- Verschraegen CF, Kantarjian HM, Hirsch-Ginsberg C, Lee MS, O′Brien S, Rios MB, et al. The breakpoint cluster region site in patients with Philadelphia chromosome-positive chronic myelogenous leukemia. Clinical, laboratory, and prognostic correlations. Cancer 1995;76:992-7.
- Inokuchi K, Inoue T, Tojo A, Futaki M, Miyake K, Yamada T, et al. A possible correlation between the type of bcr-abl hybrid messenger RNA and platelet count in Philadelphia-positive chronic myelogenous leukemia. Blood 1991;78:3125-7.
- Balatzenko G, Vundinti BR, Margarita G. Correlation between the type of bcr-abl transcripts and blood cell counts in chronic myeloid leukemia-A possible influence of mdr1 gene expression. Hematol Rep 2011;3:e3.
- Goh HG, Hwang JY, Kim SH, Lee YH, Kim YL, Kim DW. Comprehensive analysis of BCR-ABL transcript types in Korean CML patients using a newly developed multiplex RT-PCR. Transl Res 2006;148:249-56.
- Iqbal Z, Manzoor F, Iqbal M, Ali S, Sheikh N, Khan M. Frequency of BCR-ABL Fusion Oncogene Splice Variants Associated with Chronic Myeloid Leukemia (CML). J Cancer Ther 2011;2:176-80.
- de Lemos JA, de Oliveira CM, Scerni AC, Bentes AQ, Beltrão AC, Bentes IR, et al. Differential molecular response of the transcripts B2A2 and B3A2 to imatinib mesylate in chronic myeloid leukemia. Genet Mol Res 2005;4:803-11.
- Rosas-Cabral A, Martínez-Mancilla M, Ayala-Sánchez M, Vela-Ojeda J, Bahena-Reséndiz P, Vadillo-Buenfil M, et al. Analysis of Bcr-abl type transcript and its relationship with platelet count in Mexican patients with chronic myeloid leukemia. Gac Med Mex 2003;139:553-9.
- Udomsakdi-Auewarakul C, U-Pratya Y, Boonmoh S, Vatanavicharn S. Detection of molecular variants of BCR-ABL gene in bone marrow and blood of patients with chronic myeloid leukemia by reverse-transcriptase polymerase chain reaction (RT-PCR). J Med Assoc Thai 2000;83:928-35.
- Martínez-Mancilla M, Gutiérrez M, de la Rosa GZ, García-Carrancá A, Gariglio P, Miranda E. Younger age and shorter chronic phase in b2a2-positive chronic myeloid leukemia adults with high white blood cell count at diagnosis. Haematologica 2002;87:666-8.
- Shepherd P, Suffolk R, Halsey J, Allan N. Analysis of molecular breakpoint and m-RNA transcripts in a prospective randomized trial of interferon in chronic myeloid leukaemia: No correlation with clinical features, cytogenetic response, duration of chronic phase, or survival. Br J Haematol 1995;89:546-54.
- Baccarani M, Cortes J, Pane F, Niederwieser D, Saglio G, Apperley J, et al. Chronic myeloid leukemia: An update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol 2009;27:6041-51.
- Baccarani M, Saglio G, Goldman J, Hochhaus A, Simonsson B, Appelbaum F, et al. Evolving concepts in the management of chronic myeloid leukemia: Recommendations from an expert panel on behalf of the European LeukemiaNet. Blood 2006;108:1809-20.


 PDF
PDF  Views
Views  Share
Share

