Role of CTCF poly(ADP-Ribosyl)ation in the regulation of apoptosis in breast cancer cells
CC BY-NC-ND 4.0 · Indian J Med Paediatr Oncol 2015; 36(01): 49-54
DOI: DOI: 10.4103/0971-5851.151784
Abstract
Introduction: CTCF is a candidate tumor suppressor gene encoding a multifunctional transcription factor. CTCF function is controlled by posttranslational modification and interaction with other proteins. Research findings suggested that CTCF function can be regulated by poly(ADP-ribosyl)ation (PARlation) and has specific anti-apoptotic function in breast cancer cells. The aim of this study is to assess the effect of CTCF-wild type (WT) and CTCF complete mutant, which is deficient of PARlation in regulating apoptosis in breast cancer cells. Materials and Methods: The effect of CTCF-WT and CTCF complete mutant was expressed in breast cancer cell-lines by DNA-mediated transfection technique monitored by enhanced green fluorescent protein fluorescence. Evaluation of apoptotic cell death was carried out with immunohistochemical staining using 4′-6′-diamino-2 phenylindole and scoring by fluorescent microscopy. Results: CTCF-WT supports survival of breast cancer cells and was observed that CTCF complete mutant interferes with the functions of the CTCF-WT and there was a considerable apoptotic cell death in the breast cancer cell lines such as MDA-MB-435, CAMA-1 and MCF-7. Conclusion: The study enlighten CTCF as a "Biological Marker" for breast cancer and the role of CTCF PARlation may be involved in breast carcinogenesis.
Keywords
4′-6′-diamino-2 phenylindole - CTCF complete mutant - CTCF-wild type - poly(ADP-ribosyl)ation - transfectionPublication History
Article published online:
12 July 2021
© 2015. Indian Society of Medical and Paediatric Oncology. This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial-License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used for commercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/.)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
Abstract
Introduction:
CTCF is a candidate tumor suppressor gene encoding a multifunctional transcription factor. CTCF function is controlled by posttranslational modification and interaction with other proteins. Research findings suggested that CTCF function can be regulated by poly(ADP-ribosyl)ation (PARlation) and has specific anti-apoptotic function in breast cancer cells. The aim of this study is to assess the effect of CTCF-wild type (WT) and CTCF complete mutant, which is deficient of PARlation in regulating apoptosis in breast cancer cells.
Materials and Methods:
The effect of CTCF-WT and CTCF complete mutant was expressed in breast cancer cell-lines by DNA-mediated transfection technique monitored by enhanced green fluorescent protein fluorescence. Evaluation of apoptotic cell death was carried out with immunohistochemical staining using 4’-6’-diamino-2 phenylindole and scoring by fluorescent microscopy.
Results:
CTCF-WT supports survival of breast cancer cells and was observed that CTCF complete mutant interferes with the functions of the CTCF-WT and there was a considerable apoptotic cell death in the breast cancer cell lines such as MDA-MB-435, CAMA-1 and MCF-7.
Conclusion:
The study enlighten CTCF as a Biological Marker for breast cancer and the role of CTCF PARlation may be involved in breast carcinogenesis.
INTRODUCTION
The novel zinc-finger transcriptional factor, human CTCF is comprised of 728 amino acids and has a molecular weight of 130 kDa shown by sodium dodecyl sulfate polyacrylamide gel electrophoresis. CTCF is a multivalent, multifunctional ubiquitously expressed, nuclear phosphoprotein with a DNA binding domain containing 11 zinc-fingers. [1,2] CTCF has been characterized as a true “multifunctional” protein because it utilizes different combinations of individual zinc fingers to form distinct complexes to mediate functions in regulation of gene expression. CTCF regulates the role as promoter activation or expression or repression, methylation dependent chromatin insulation and genomic imprinting. [3] CTCF has been found to undergo posttranslational modification by phosphorylation. CTCF can be phosphorylated at serine sites and it does not affect CTCF nuclear localization. [4]
Tumor suppressor genes are responsible for restraining cell growth and alteration to their structure and can lead to abnormal functions which, in turn result in cancer. CTCF is considered to be a candidate tumor suppressor gene as it exhibits classical features that characterize tumor suppressor. The functional significance tumor specific CTCF zinc finger mutations have been identified in different types of tumors which, includes breast cancer. The CTCF biological function confirms the link to epigenetic mechanisms, together with established features of a tumor suppressor suggesting that CTCF has an important role in tumorigenesis. Breast tumor specific mutations within the CTCF zinc finger domain has been discovered and found to be a single nucleotide substitution resulting in a mis-sense codon. [5]
Poly(ADP-ribosyl)ation (PARlation) is a posttranslational modification of the amino-acids such as glutamate and aspartate of nuclear/acceptor proteins. Major function of poly (ADP-ribose) polymerase-1 (PARP-1) is as a survival factor involved in recovery from DNA damage in cells and there is evidence that it is implicated in the maintenance of the genomic stability of cells and detects specifically DNA strand breaks generated by genotoxic agents. [6] It is suggested that binding of CTCF was dependent upon functional CTCF target sites and importantly, upon CTCF modification by PARlation and has an impact on the biological function. [7] Apoptosis is a form of programmed cell death in which, a controlled sequence of events leads to the elimination of cells without realizing harmful substances in to the surrounding area. The process of programmed cell death plays an important role at different stages of carcinogenesis and generally cancer cells are highly sensitized to the apoptotic signals. [8]
In the present study, we investigated the role of CTCF PARlation in the regulation of apoptosis in breast cancer cells. To achieve this goal the effect of CTCF-wild type (CTCF-WT) and CTCF complete mutant (mutant 4) was transiently transfected into the human breast cancer cells lines such as MDA-MB-435, CAMA-1, and MCF-7. The regulation of apoptosis has been monitored using sodium butyrate (NaB) due to its nature to induce apoptosis. The experiment has also been compared with that of the nonbreast cancer cell line human embryonic kidney 293T (HEK-293T).
In this study, we hypothesize the effect of CTCF complete mutant which, is deficient in PARlation as individually and also with NaB to regulate the apoptosis in breast cancer cell-lines because CTCF complete mutant has the attribute which, is considered to interfere with the functions of the CTCF-WT on the contrary which, promotes the breast cancer cell survival.
MATERIALS AND METHODS
Cell lines
The following cell lines were used as experiment model for the research: 293T-human kidney cell line: Adherent, epithelial cells were established from human primary embryonic kidney transformed by adenovirus type 5 (AD 5) also known as HEK-293T (ATCC, CRL-11268). 293T is a highly transfectable derivative of the 293 cell line and the growing medium is Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, USA) supplemented with 10% (v/v) heat inactivated fetal bovine serum (FBS) (Sigma, USA) and 0.5% (v/v) heat stable antibiotic gentamycin (Gibco, BRL).
Breast carcinoma cell lines
- ZR-75-1: This cell line composed of adherent epithelial estrogen dependent breast cancer cells (ATCC, CRL-1500). They have a more circular structure than other breast cancer cell lines and tend to grow as monolayer with mosaic shaped clusters in culture.
- MCF-7: These are epithelial cells derived from a human breast carcinoma (ATCC, HTB-22D). They tend to grow into clusters which, leads to mosaic.
- CAMA-1: Isolated as adherent patches of epithelial cells from malignant pleural effusion derived from a human breast carcinoma (ATCC, HTB-21). It grows as compost, multilayered colonies in culture.
- MDA-MB-435: This cell line was derived from pleural effusion and cells appear morphologically epithelial like and are considered to be highly metastatic (ATCC, HTB-129). All the breast carcinoma cell lines have been grown in Rosewell Park Memorial Institute 1640 (RPMI) (Invirogen, USA) with ultra-glutamine 1: Supplemented with 10% (v/v) heat activated FBS (Sigma, USA) and 0.5% (v/v) antibiotic gentamycin (Gibco, BRL). Cell lines were maintained at 37°C in an atmosphere of 5% CO2.
Plasmid DNA/vector
Plasmid enhanced green fluorescent protein (pEGFP)-C1 plasmid pEGFP-C1 encodes GFP with 238 amino acid residues originally cloned from the jelly fish Aequorea victoria. It is being frequently used in order to assess the transfection efficiency due to its ability to emit a fluorescent green light on exposure to blue or ultraviolet (UV) light. The source of pEGFP-C1 plasmid is from Clonetech, USA. pEGFP-CTCF-WT and pEGFP-CTCF-M4 was provided from the laboratory of Professor Elena Kelnova.
Methods
All breast carcinoma cell lines were cultured in RPMI 1640 while the 293T kidney cell line was cultured in DMEM medium. Both medium were supplemented with 10% (v/v) FBS and 0.5% (v/v) gentamycin.
Cell passaging technique
The cell cultures were grown in 150 ml sterile flasks with approximately 15 ml appropriate medium and incubated at 37°C under controlled conditions of 5.0% CO2. The cell confluence and medium conditions were checked regularly and splitting was carried out when cells become sub-confluent so as to avoid cells leaving the cell cycle and thus causing cell death.
On every 3rd day a 1:2 splitting was required where half of the cells were removed into the aspirating flask and the same volume was replaced by free fresh medium and made up to 15 ml.
Transfection
Principle
Gene cloning or introducing a gene of interest into different cell types in order to observe the newly introduced gene and its effect on expression with regard to that of other genes.
For our experiment procedure we have carried out “transient transfection” using calcium phosphate. Before transfection we need to ensure that the well plate for the experiment was checked for 70% confluence of cells. An hour prior to transfection, the medium covering the cells was removed and replaced with prewarmed antibiotic free medium DMEM by Gibco supplemented with 10% (v/v) heat inactivated FBS (Sigma, USA). The plates were then returned to the incubator, while preparing the DNA. The total amount of DNA tansfected was 2 μg (4 μl). In each set of experiment a sample of empty expression vector was made. The appropriate volume of vectors was pipette into 1.5 ml eppendorf tubes and the volume was made up to 45 μl using sterile water. Then, 5 μl of calcium chloride (CaCl2) (2.5 mM) was added to the DNA and the tube containing DNA was incubated in the ice for 7 min. The incubated DNA-CaCl2 was added drop wise into separate fresh eppendorf tube containing 50 μl of N,N-Bis (2-hydroxyethyl)-2-aminoethanesulfonic acid (50 mM 2xBES (BES), 280 mM Nacl, 1.5 mM Na2 HPO4, pH 6.9) in room temperature or 2xHBS (280 mM Nacl, 1.5 mM Na2 HPO4, 50 mM Hepes, pH 7) as in the case of HEK-293T and MCF-7 and vortexing slowly to ensure proper mixing of the entire volume. The entire volume of 100 μl of DNA solution was incubated at room temperature for 20 min and then added drop-wise onto the surface of the medium covering the cells and slightly swirling the plate to distribute evenly. The plated cells were returned overnight to the incubator. After 24 h of incubation the transfection medium was removed and replaced with fresh prewarmed DMEM complete medium for HEK-293T cells and RPMI complete medium for breast cancer cell lines.
The absence of antibiotic on the transfection medium was necessary because CaCl2 would react with antibiotic (gentamycin) resulting in the inhibition of the transfection. RPMI was not an appropriate transfection medium as its positive charge could cause the formation of dense precipitation. Two micro gram pEGFP, 2 μg pEGFP-CTCF-WT, 2 μg pEGFP-CTCF-M4 are the combination of vectors used for the present study.
Twenty-four hour after transfection, the cells were examined under a fluorescent microscope to visualize the uptake of enhanced green fluorescent protein (EGFP) (fluoresce bright green color). Three random fields were used per well and the transfection efficiency obtained by counting the number of green fluoresced cells as a percentage of total cells per field of view. In these experiments an IMT-2 a 35 mm Phase Contrast Olympus Fluorescent Microscope was used.
Chemical induction of apoptosis
Apoptotic cell death was chemically induced by NaB. The molecular mechanism of NaB action has been well understood. [9] NaB also increases the transfection efficiency and expression of transient transfection. [10] The breast cancer and nonbreast cancer cell lines used for comparison in this experiment were transiently transfected and after 24 h they were treated with 5 mM NaB.
Fixing and 4’-6’-diamino-2 phenylindole staining
Principle
4΄-6΄-diamino-2 phenylindole (DAPI) known to form fluorescent complexes with natural double stranded DNA, showing fluorescence specificity for AT, AU and IC (Ion Channel) clusters.
The medium in the well plate was carefully removed without disturbing the cover slip using aspirating pipette. On removal of medium per well 600 μl of phosphate buffered saline (1xPBS) was used for washing the cells. The well plate were returned to the incubator for drying 10-15 min by ensuring the cover slip with cells were dried properly for further procedure. 500 μl of 4% formaldehyde PBS (219.20 μls formaldehyde in 1.78 ml PBS) was added to each well and the well plate were covered with silver foil in order to prevent the external light hindering formaldehyde reaction being light sensitive. The well plate was kept for 30 min incubation for formaldehyde reaction and washed again with 500 μl PBS by gently shaking the well plate for 10 min. The cover slip where the cells adhered on the well was removed gently and mounted on a slide. 30 μl of DAPI (1 μg of DAPI: 400 μl PBS) was added to the cover slip with coverage on all parts of the cover slip and kept undisturbed for 15 min. [11] The cover slip from the slide was replaced in the respective well on the well plate and washed with 500 μl PBS for 10 min. After washing the cover slip was removed and air dried by placing it on a tissue paper. On drying the cover slip was mounted upside down on a microscopic slide using 1-2 drops of fluorescent mounting medium (DAKO cytomation). The fixed cover slip on the microscopic slide was left undisturbed for few minutes to air dry and placed in slide holders for microscopic observations.
Assessment of apoptosis
24-48 h after treatment with NaB, the cells were examined under fluorescence microscopy to visualize and calculate the uptake of EGFP (fluoresce bright green color) for determining the transfection efficiency in relation to apoptosis. The morphology of the cells nuclei was observed using the same microscope at an excitation wavelength 350 nm (UV light) for apoptosis. Nuclei are considered to have the normal phenotype when glowing bright and homogenously. An apoptotic nucleus was identified by the condensed chromatin gathering at the periphery of the nuclear membrane or a total fragmented morphology of nuclear bodies. More than 300 cells were counted and the percentage of apoptotic nuclei determined. In these experiments Olympus BX41 Upright Microscope was used.
RESULTS
Studies on HEK-293T cell line where analyzed and measured on 24, 48, 72 and 96 h after plating. It was observed an exponential increase in the number of HEK-293T cancer cell multiplication which has been proportional to the hours of incubation. The transfection studies with different ρEGFP concentration showed the higher transfection efficiency was achieved with 0.75 μg than that of 0.25 μg.
Transfection studies have also been carried out on the breast cancer cell lines such as ZR-75-1 and MDA-MB-435 using Plasmid DNA EGFP (as internal marker), CTCF-WT and C TCF complete mutant. Two different plasmid concentrations such as 1 μg and 2 μg were used in the study. Cells were observed with IMT-2 35 mm phase contrast Olympus Fluorescent Microscopy after 24, 48 and 72 h of transfection and recorded the scores for the number of cells which were transfected. The recorded values in the graph indicate a very low rate of transfection with no increase in the rate of transfection even after prolonged period of incubation in the ZR-75-1 cell line [Figure 1] compared with that of MDA-MB-435 breast cancer cell line [Figure 2]. It was observed that MDA-MB-435 cell line looks to be promising for further carryout on experiments than ZR-75-1.
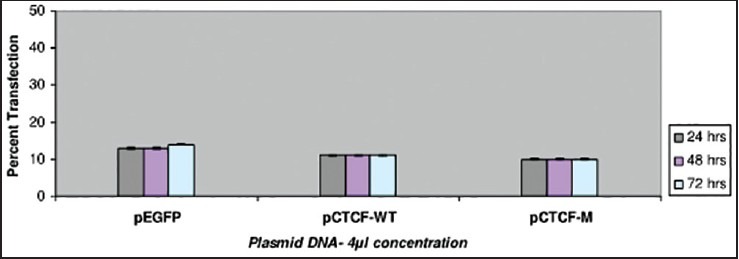
| Fig. 1 Bar graph showing the transfection studies with ZR-75 breast cancer cell-line. The recorded values in the graph indicate a very low rate of transfection with no increase in the rate being observed even after prolonged incubation
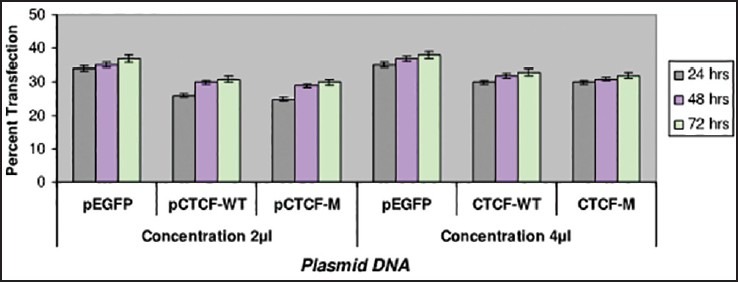
| Fig. 2 Bar graph showing the transfection studies with MDAMB- 435 breast cancer cell-line. The recorded values showed promising efficiency on MDA-MB-435 cell line in comparison with ZR-75
Studies have been carried out with different concentrations of NaB at 1 mM, 5 mM and 10 mM. The breast cancer cell line MDA-MB-435 and was observed to have the maximum apoptotic cell death of 37% at 72 h on 10 mM NaB [Figure 3] in comparison with the CAMA-1 and MCF-7 cell lines.
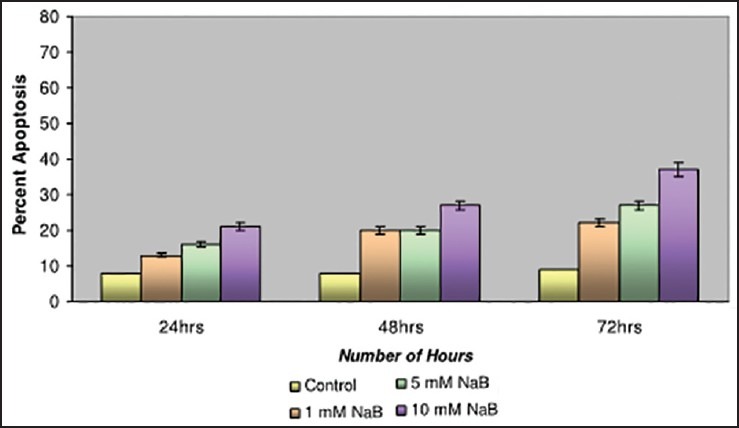
| Fig. 3 Bar graph shows the studies with Sodium butyrate on MDAMB- 435 cell line. The apoptotic cell death was recorded as 37% at 72 h
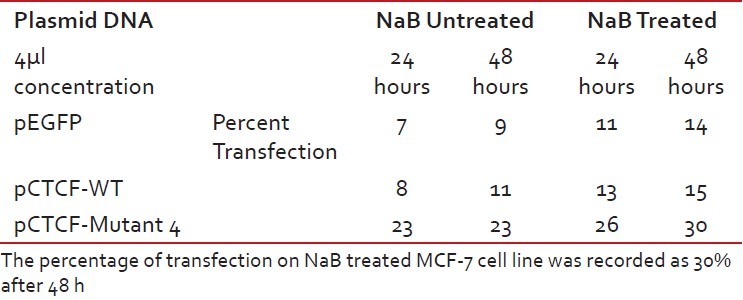
Finally, studies on the effect of transfection and NaB have been carried out on the above mentioned breast cancer cell lines such as CAMA-1, MCF-7 and MDA-MB-435. The results have been tabulated. From the readings it was observed that the efficiency of transfection on NaB treated complete mutant 4 MDA-MB-435 cell line showed the maximum at 48 h as 77% [Table 1], followed by CAMA-1 [Table 2] showing 56% and MCF-7 with 30% at 48 h, respectively [Table 3]. The results were also recorded for the apoptosis on all the above mentioned transfected and NaB treated cell lines using BX41 Fluorescent Microscopy. The highest recorded apoptotic cell death was observed in MDA-MB-435 cell line showing 51% at 24 h and 73% in 48 h with complete mutant 4 [Figure 4] in comparison to that of CAMA-1 [Figure 5] and MCF-7 [Figure 6].
Table 1
Transfection efficiency in MDA-MB 435 breast cancer cell line
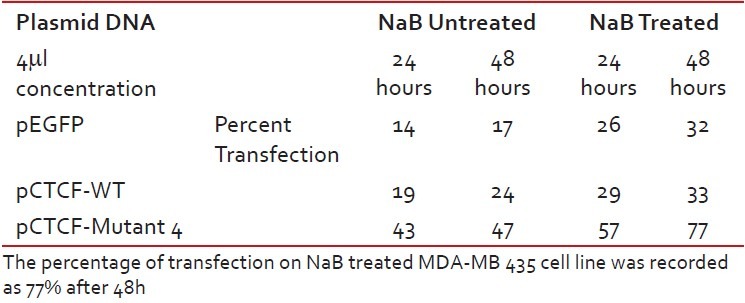
Table 2
Transfection efficiency in CAMA-1 breast cancer cell line
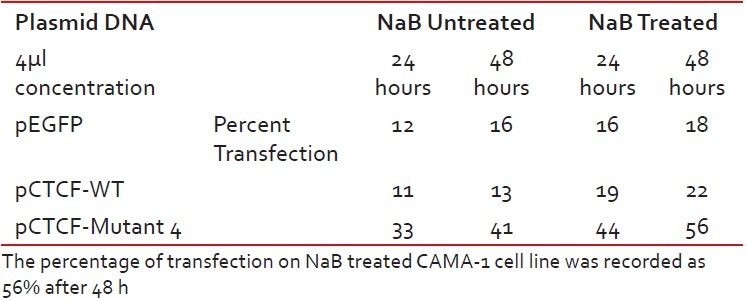
Table 3
Transfection efficiency in MCF-7 breast cancer cell line
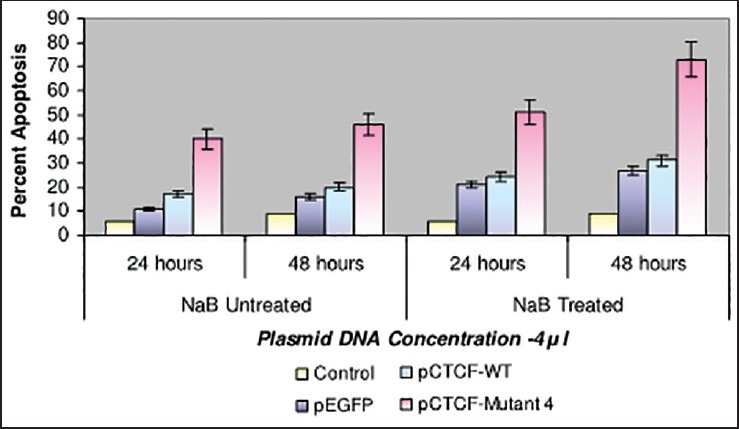
| Fig. 4 Bar graph shows the percent apoptosis on the sodium butyrate treated and transfected MDA-MB-435 cell line. The percent efficiency has been recorded as 51% at 24 h and 73% at 48 h
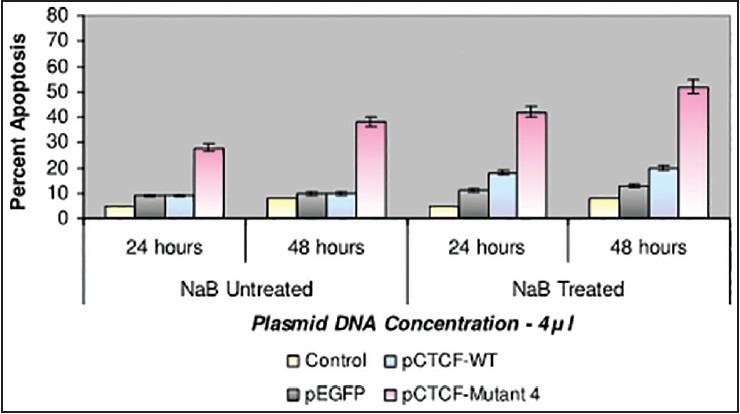
| Fig. 5 Bar graph shows the percent apoptosis on the sodium butyrate treated and transfected CAMA-1 cell line. The percent efficiency has been recorded as 42% at 24 h and 52% at 48 h
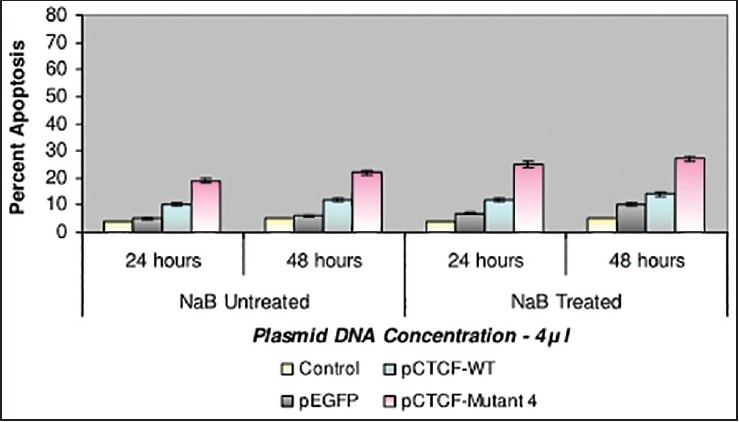
| Fig. 6 Bar graph shows the percent apoptosis on the sodium butyrate treated and transfected MCF-7 cell line. The percent efficiency has been recorded as 25% at 24 h and 27% at 48 h
DISCUSSION
This study aim to investigate the role of CTCF-WT and CTCF complete mutant deficient of PARlation in regulating the breast cancer cells. The results showed that CTCF complete mutant along with NaB has a remarkable percent of apoptosis in MDA-MB-435 breast cancer cell line showing 73%. The apoptosis percent was also considerable in the other two breast cancer cell lines such as CAMA-1 and MCF-7. The analysis also revealed CTCF WT showing a very low percent of apoptosis compared to CTCF complete mutant on all the breast cancer cell lines. This may be a significant finding because the analysis indicates a positive correlation between CTCF complete mutant and apoptotic cell death.
Generally accepted criterion is that cancer cells are highly sensitized to apoptotic signals, and normal cells are more resistant to apoptotic challenges. [12] Using both transient transfection approach with CTCF-WT and CTCF complete mutant and also an inducible approach using NaB the endogenous levels of CTCF PARlation has led to apoptotic cell death in the breast cancer cell lines than CTCF-WT. These results match to the findings of Docquier et al. for CTCF expression and apoptotic cell death. [13] CTCF a candidate tumor suppressor is involved in regulation of genes controlling proliferation. This statement and findings seem to conflict with the findings of Docquier et al. where CTCF being involved in the regulation of apoptosis and proliferation by having “dual signal”. Still their findings became the key reference on this research and the results support the view that CTCF-WT having higher expression of CTCF PARlation showing very insignificant apoptotic cell death and on the other hand CTCF complete mutant shows a very remarkable apoptotic cell death.
The percent effect of apoptosis on both CTCF WT and CTCF complete mutant has been to some extent strengthened due to NaB. These results are in agreement with the findings of Chopin et al. who has reported butyrate induced cell cycle arrest in G1 and apoptosis in MCF-7. T47-D, and BT-20 cells, and arrest in GA/M in MDA-MB-231 cells where butyrate has been demonstrated as a potent growth inhibitor.
Research findings by Guastafierro et al., suggested that CTCF has been involved in the cross talk between PARlation and DNA methylation and underscored the importance of rapid reversal of PARP activity, as DNA methylation pattern getting responsible for an important epigenetic code. [14] CTCF, a CCCTC binding factor, has a diverse role in DNA replication, insulation, and genome regulation, but remains unclear whether subsets of CTCF binding site play a functional role in establishing and maintain the topological domain. [15]
CONCLUSION
Present study demonstrates CTCF complete mutant has an appreciable percent effect in regulating the apoptosis on the breast cancer cell lines in comparison with that of the CTCF-WT. This study enlightens CTCF as a “Biological Marker” for breast cancer and shows that the role of CTCF PARlation may be involved in breast carcinogenesis. The future proceedings has to envisage onto the finer details of all molecular mechanism(s) corresponding to CTCF binding factor and PARlation.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES

| Fig. 1 Bar graph showing the transfection studies with ZR-75 breast cancer cell-line. The recorded values in the graph indicate a very low rate of transfection with no increase in the rate being observed even after prolonged incubation

| Fig. 2 Bar graph showing the transfection studies with MDAMB- 435 breast cancer cell-line. The recorded values showed promising efficiency on MDA-MB-435 cell line in comparison with ZR-75

| Fig. 3 Bar graph shows the studies with Sodium butyrate on MDAMB- 435 cell line. The apoptotic cell death was recorded as 37% at 72 h

| Fig. 4 Bar graph shows the percent apoptosis on the sodium butyrate treated and transfected MDA-MB-435 cell line. The percent efficiency has been recorded as 51% at 24 h and 73% at 48 h

| Fig. 5 Bar graph shows the percent apoptosis on the sodium butyrate treated and transfected CAMA-1 cell line. The percent efficiency has been recorded as 42% at 24 h and 52% at 48 h

| Fig. 6 Bar graph shows the percent apoptosis on the sodium butyrate treated and transfected MCF-7 cell line. The percent efficiency has been recorded as 25% at 24 h and 27% at 48 h


 PDF
PDF  Views
Views  Share
Share

