Deep Learning Based Classification of Cervical Cancer Stages Using Transfer Learning Models
CC BY 4.0 · Indian J Med Paediatr Oncol 2025; 46(05): 480-488
DOI: DOI: 10.1055/s-0045-1809907
Abstract
Introduction
Cervical cancer is one of the leading causes of mortality among women, emphasizing the need for accurate diagnostic methods particularly in developing countries where access to regular screening is limited. Early detection and accurate classification of cervical cancer stages are crucial for effective treatment and improved survival rates.
Objectives
This study explores the potential of deep learning based convolutional neural networks (CNNs) for classifying cervical cytological images from the Sipahan Kanker Metadata (SIPaKMeD) dataset.
Materials and Methods
The SIPaKMeD dataset originally containing 4,049 images is augmented to 24,294 images to enhance model generalization. We employed VGG-16, EfficientNet-B7, and CapsNet CNN models using transfer learning with ImageNet pretrained weights to improve classification accuracy.
Results
The experimental results show that EfficientNet-B7 achieved the average highest classification accuracy of 91.34%, outperforming VGG-16 (86.5%) and CapsNet (81.34%). Evaluation metrics such as precision, recall, and F1-score further validate the robustness of EfficientNet-B7 in distinguishing between different cervical cancer stages. After testing with various hyperparameters, EfficientNet-B7 minimizes misclassification errors and is able to categorize data more accurately compared to other CNN models.
Conclusion
These findings highlight the potential of deep learning CNNs for automated cervical cancer diagnosis, aiding doctors in clinical decision-making to classify medical images and diagnose diseases. Consequently, diagnostic accuracy improves, facilitating more effective treatment planning in the healthcare sector.
Keywords
female - uterine cervical neoplasms - early detection of cancer - deep neural networks - pathologistsData Availability Statement
Datasets associated with this article are available at (https://universe.roboflow.com/ik-zu-quan-o9tdm/sipakmed-ioflq).
Patient's Consent
Patient consent was waived as the data used were anonymized and obtained from publicly available sources.
Supplementary MaterialPublication History
Article published online:
03 July 2025
© 2025. The Author(s). This is an open access article published by Thieme under the terms of the Creative Commons Attribution License, permitting unrestricted use, distribution, and reproduction so long as the original work is properly cited. (https://creativecommons.org/licenses/by/4.0/)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
- Deep learning radiomics of multimodal ultrasound for classifying metastatic cervical lymphadenopathy into primary cancer sites: a feasibility studyYangyang Zhu, Ultraschall in der Medizin - European Journal of Ultrasound
- Artificial Intelligence for early identification of Amyloid positivity in transgenic Alzheimer miceF. Eckenweber, Nuklearmedizin, 2022
- Deep-learning-based clinical decision support system for gastric neoplasms in real-time endoscopy: Development and validation studyEun Jeong Gong, Endoscopy
- Advancing Medical Imaging Informatics by Deep Learning-Based Domain AdaptationAnirudh Choudhary, Yearbook of Medical Informatics
- Automated classification of gastric neoplasms in endoscopic images using a convolutional neural networkBum-Joo Cho, Endoscopy, 2019
- Assessments of Data-Driven Deep Learning Models on One-Month Predictions of Pan-Arctic Sea Ice Thickness<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- Software Skills Identification: A Multi-Class Classification on Source Code Using Machine Learning<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- Pancancer outcome prediction via a unified weakly supervised deep learning model<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- Deep Learning Classification of Epileptic Magnetoencephalogram<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- A data-driven method for microgrid bidding optimization in electricity market<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
Abstract
Introduction
Cervical cancer is one of the leading causes of mortality among women, emphasizing the need for accurate diagnostic methods particularly in developing countries where access to regular screening is limited. Early detection and accurate classification of cervical cancer stages are crucial for effective treatment and improved survival rates.
Objectives
This study explores the potential of deep learning based convolutional neural networks (CNNs) for classifying cervical cytological images from the Sipahan Kanker Metadata (SIPaKMeD) dataset.
Materials and Methods
The SIPaKMeD dataset originally containing 4,049 images is augmented to 24,294 images to enhance model generalization. We employed VGG-16, EfficientNet-B7, and CapsNet CNN models using transfer learning with ImageNet pretrained weights to improve classification accuracy.
Results
The experimental results show that EfficientNet-B7 achieved the average highest classification accuracy of 91.34%, outperforming VGG-16 (86.5%) and CapsNet (81.34%). Evaluation metrics such as precision, recall, and F1-score further validate the robustness of EfficientNet-B7 in distinguishing between different cervical cancer stages. After testing with various hyperparameters, EfficientNet-B7 minimizes misclassification errors and is able to categorize data more accurately compared to other CNN models.
Conclusion
These findings highlight the potential of deep learning CNNs for automated cervical cancer diagnosis, aiding doctors in clinical decision-making to classify medical images and diagnose diseases. Consequently, diagnostic accuracy improves, facilitating more effective treatment planning in the healthcare sector.
Keywords
female - uterine cervical neoplasms - early detection of cancer - deep neural networks - pathologistsIntroduction
Cervical cancer is one of the most prevalent cancers among women worldwide particularly in low and middle income countries, where access to screening and early detection is limited.[1] It arises from the cervix, the lower part of the uterus that connects to the vagina. The primary cause of cervical cancer is persistent infection with high risk strains of the human papillomavirus,[2] a sexually transmitted virus that can cause changes in the cells of the cervix. If these changes are left untreated, they can lead to cervical dysplasia and eventually invasive cancer.[3] Cervical cancer progresses through various stages each reflecting the severity of the disease and its potential to affect surrounding tissues. The detection of these stages plays a critical role in guiding treatment decisions, predicting patient outcomes, and improving survival rates. Staging helps to determine whether cancer is localized to the cervix or has spread to nearby tissues or distant organs. Early detection through effective screening and accurate staging is essential for early intervention, which is associated with higher survival rates and less invasive treatments.[4]
The most common system used for staging cervical cancer is the FIGO (International Federation of Gynecology and Obstetrics) system, which divides the disease into several stages based on the size of the tumor, extent of invasion, and involvement of lymph nodes or distant organs.[5] However, precancerous changes or low-grade lesions (cervical intraepithelial neoplasia) can be observed before cancer develops. Early identification of these stages can significantly reduce the risk of progression to invasive cancer. The accurate detection and grading of cervical lesions are critical for determining the appropriate treatment options.[6] The earlier the cancer is detected and staged the more effective the treatment, leading to higher survival rates and better quality of life for patients.
Recent advancements in machine learning (ML) and artificial intelligence have opened new possibilities for the automatic detection and classification of cervical cancer stages. Medical imaging using colposcopy, Pap smears, and cervical biopsies combined with deep learning algorithms can be used to detect and classify cervical lesions with high accuracy.[6] [7] [8] [9] One promising approach is the use of novel deep learning architecture that can capture spatial hierarchies in images. Convolutional neural network (CNN) ability to model part-whole relationships makes it particularly effective for tasks like cervical pathology classification, where accurate identification of abnormalities and staging is critical. CNN models can be trained on large annotated datasets to automatically classify cervical lesions into stages based on severity, reducing the reliability of manual interpretations and increasing diagnostic accuracy. The cutting-edge technologies such as computer vision, ML, and digital image processing that have been shown to be successful in detecting cervical cancer are reviewed and summarized in [Table 1].
|
First author, reference, and year |
Image dataset |
Feature |
Preprocessing |
Segmentation |
Classifier |
Results |
|---|---|---|---|---|---|---|
|
Bora et al[3] 2017 |
Dr. B. Borooah Cancer Institute, India, Herlev and Ayursundra Healthcare Pvt. Ltd |
Shape, texture, and color |
Bit plane slicing, median filter, LL filter |
DWT with MSER |
Ensemble classifier |
Overall classification accuracy of 98.11% and precision of 98.38% at smear level and 99.01% at cell level with ensemble classifier. Using the Herlev database an accuracy of 96.51% (2 class) and 91.71% (3 class) |
|
Yakkundimath et al[4] 2022 |
Herlev |
Colors and textures |
SVM, RF, and ANN |
Overall classification accuracy of 93.44% |
||
|
Chen et al[6] 2023 |
Private dataset |
ROI is extracted |
EfficientNet-b0 and GRU |
EfficientNet |
Overall classification accuracy of 91.18% |
|
|
Cheng et al[7] 2021 |
Private |
Resolution of 0.243 μm per pixel |
ResNet-152 and Inception-ResNet-v2 |
– |
ResNet50 |
Overall classification accuracy of 0.96 |
|
Liu et al[8] 2020 |
Private |
Normalization |
DCNN with ETS |
– |
VGG-16 |
Overall classification accuracy of 98.07% |
|
Sreedevi et al[9] 2012 |
Herlev |
Morphological |
Normalization, scaling, and color conversion |
Iterative thresholding |
Area of nucleus |
Specificity of 90% and sensitivity of 100% |
|
Genctav et al[16] 2011 |
Herlev and Hacettepe |
Nucleus such as area |
Preprocessed |
Automatic thresholding and multi-scale hierarchical |
SVM, Bayesian, decision tree |
Overall classification rate of 96.71% |
|
Talukdar et al[17] 2013 |
Color image |
Colorimetric and textural, densitometric, morphometric |
Otsu's method with adaptive histogram equalization |
Chaos theory corresponding to RGB value |
Pixel-level classification and shape analysis |
Minimal data loss preserving color images |
|
Lu et al[18] 2013 |
Synthetic image |
Cell, nucleus, cytoplasm |
EDF algorithm with complete discrete wavelet transform, filter |
Scene segmentation |
MSER |
Jac-card index of > 0.8, precision of 0.69, and recall of 0.90 is obtained |
|
Chankong et al[19] 2014 |
Herlev ERUDIT and LCH |
Cytoplasm, nucleus, and background |
Preprocessed |
FCM |
FCM |
Overall classification rate of 93.78% for 7-class and 99.27% for 2-class. |
|
Kumar et al[20] 2015 |
Histology |
Morphological |
Adaptive histogram equalization |
K-means |
SVM, K-NN, random forest, and fuzzy KNN |
Accuracy of 92%, specificity of 94%, and sensitivity of 81% |
|
Sharma et al[21] 2016 |
Fortis Hospital, India |
Morphological features |
Histogram equalization and Gaussian filter |
Edge detection and min.–max. methods |
K-NN |
Overall classification accuracy of 82.9% with fivefold cross-validation |
|
Su et al[22] 2016 |
Epithelial cells from liquid-based cytology slides |
Morphological and texture |
Median filter and Histogram equalization |
Adaptive threshold segmentation |
C4.5 and logical regression |
Overall classification accuracy of 92.7% with the C4.5 classifier 93.2%, with the LR classifier, and 95.6% with the integrated classifier |
|
Ashok et al[23] 2016 |
Rajah Muthiah Medical College, India |
Texture and shape |
Image resizing, filters, and Grayscale image conversation |
Multi-thresholding |
SVM |
Overall classification accuracy of 98.5%, sensitivity of 98%, and specificity of 97.5% |
|
Bhowmik et al[24] 2018 |
AGMC-TU |
Shape |
Anisotropic diffusion |
Mean-shift, FCM, region-growing, K-means clustering |
SVM-Linear(SVM-L) |
Overall classification accuracy of 92.83% with SVM linear (SVM-L) and 97.65% with a discriminative feature set |
|
William et al[25] 2019 |
MRRH, Uganda |
Morphological |
Contrast local adaptive histogram equalization |
TWS |
FCM |
Overall classification accuracy of 98.8%, sensitivity of 99.28% and specificity of 97.47% |
|
Zhang et al[26] 2017 |
Herlev |
Deep hierarchical |
Preprocessed |
Sampling |
ConvNet CNN |
Overall classification accuracy of 98.3%, area under the curve of 0.9, and specificity of 98.3% |
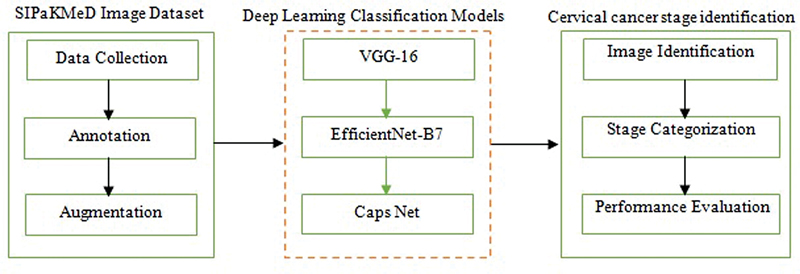
| Fig 1 : Schematic overview of the proposed method.
Image Dataset
The SIPaKMeD[10] dataset contains a total of labeled 4,049 images distributed across five classes, namely, superficial-intermediate (831 images), parabasal (787 images), koilocytotic (825 images), dyskeratotic (813 images), and metaplastic (793 images). Each image is an RGB file with a resolution of 128 × 128 pixels, ensuring uniformity across the dataset for preprocessing and algorithmic analysis. The expert annotations provided for all images serve as reliable ground truth, enabling the development and benchmarking of automated cytological image analysis methods. Further, the SIPaKMeD dataset is categorized into stages 0 to 4, reflecting the progression of cervical pathology mapped to the classes such as stage 0 (normal/benign), stage 1 (benign immature), stage 2 (low-grade lesions), stage 3 (high-grade lesions), and stage 4 (advanced metaplastic/repairing cells). This classification correlates with the progression of cellular changes observed in cervical pathology and is useful for applying SIPaKMeD in diagnostic research and model development. Some of the sample images of cervical cancer used for categorization based on stages are shown in [Fig. 2]. Based on the stages and the number of images in each class of the SIPaKMeD dataset, the distribution of images across the stages is given in [Supplementary Table 1] (available in the online version only).

| fig 2:Stages of cervical cancer.
Image Augmentation
Image augmentation is essential when working with CNNs to enhance the diversity of the training dataset and improve the model's robustness to increase the potential imbalance between classes. The number of images in the SIPaKMeD dataset is substantially increased through classical augmentation techniques such as rotation, flipping, scaling, brightness adjustment, and translation.[11] For the SIPaKMeD dataset originally containing 4,049 images, a classical augmentation technique is applied. Each image undergoes five classical augmentations, and the dataset is expanded by a factor of five resulting in a total of 24,294 images. The number of images after applying the augmentation technique is given in [Supplementary Table 2] (available in the online version only).
Classifier
Convolutional Neural Network
CNN models such as VGG-16,[12] EfficientNet B-7,[13] and CapsNet[14] are employed for the classification of 24,294 images derived from the SIPaKMeD dataset, which contains cytological images categorized into five classes. The architecture consists of an input layer, convolution layers, rectified linear unit (ReLU) activation layers, max-pooling layers, and a fully connected output layer tailored to multi-class classification. The input layer accepts image data in a structured format, resized to standard dimensions suitable for the model. The convolution layer extracts local features such as edges and textures, using learnable filters or kernels, while strides and padding control the resolution of the resulting feature maps. The ReLU layer introduces nonlinearity by applying the activation function f(x) = max(0,x), enabling the network to model complex relationships. The max.-pooling layer reduces the spatial dimensions of feature maps by retaining the most significant values in a defined window, minimizing computational complexity and overfitting. Finally, the fully connected layer connects the extracted features to the output layer, where activation functions like softmax or sigmoid compute class probabilities. This hierarchical architecture allows CNNs to process images effectively for tasks such as detection and classification. The Adam optimizer and categorical cross-entropy loss function are employed for optimization.[11]
Transfer Learning
The study leverages a dataset of 24,294 images derived from the SIPaKMeD dataset for the classification of cervical cytology images. Transfer learning is applied to the pretrained models such as VGG-16, EfficientNet-B7, and CapsNet CNN models. The application of transfer learning involves utilizing the knowledge embedded in pretrained models to adapt them for specific tasks with limited data. An ImageNet pretrained was utilized in this study with weights derived from training on the ImageNet dataset.[8] The ImageNet dataset comprising approximately 1.2 million images across 1,000 classes, provides a robust foundation for transfer learning. These pretrained weights are fine-tuned for the classification task on an augmented SIPaKMeD dataset containing 24,294 images. ImageNet pretrained weights are employed to initialize all the layers of the VGG-16, EfficientNet-B7, and CapsNet models. The networks are further tuned using the stochastic gradient descent (SGD) algorithm to minimize the loss function and ensure convergence.[11]
Classification
The CNN is trained, validated, and tested on the 24,294 images from the SIPaKMeD dataset. To evaluate the model's ability to generalize, the dataset is split into three subsets such as training, testing, and validation. Approximately 70% of the data (17,005 images) is used for training the model. Around 15% of the data (3,644 images) is used for validation during training.[15] This set is used to tune hyperparameters listed in [Supplementary Table 3] (available in the online version only) and assess the model's performance after each epoch. The remaining 15% of the dataset (3,645 images) is used to assess the final model's accuracy and performance after training.
The classification efficiency of pretrained VGG-16, EfficientNet-B7, and CapsNet CNN models is computed using Expressions (1) and (2). Metrics such as accuracy, precision, recall, and F1-score given in the Expressions (3) to (5) are used to check how well the model is generalizing to unseen data. The CNN classification methodology based on transfer learning adopted is given in Algorithm 1.[11] This algorithm outlines the process of classifying the SIPaKMeD dataset using pretrained VGG-16, EfficientNet-B7, and CapsNet models.
| Fig 1 :
| Figure 2:
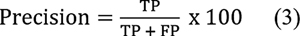
| Figure 3:
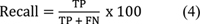
| Figure 4:
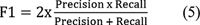
| Figure 5:
Algorithm 1
Description
The augmented SIPaKMeD dataset is first fed as input into the CNN model. The base and upper layers of the CNN are responsible for extracting and computing significant features required for classification. The algorithm processes these features to classify the images and finally produces a consolidated confusion matrix to evaluate the classification performance on the test dataset.
The following parameters are used in this algorithm.
ε: Number of epochs, ξ: Iteration step, β: batch size (number of training examples in each mini-batch); η: CNN learning rate; w: Pretrained ImageNet weights, n: number of training examples in each iteration, P (xi = k): probability of input xi being classified as the predicted class k, k: classes index.
// Input: Xtrain: SIPaKMeD training dataset, Xtest: SIPaKMeD testing dataset.
// Output: Consolidated assessment metrics corresponding to the classification performance.
Step 1. Randomly select samples for training, validation, and testing. The dataset is split into training (Xtrain), testing (Xtest), and validation sets.
Step 2. Configure the base layers of the CNN models (VGG-16, EfficientNet-B7, CapsNet), including the input layer, hidden layers, and output layer.
Step 3. Configure the upper layers of the models, that is, convolution, pooling, flattening, and dropout layers.
Step 4. Define parameters: ε, β, η.
Step 5: Initialize the model with pretrained ImageNet weights (w1, w2, …, wn) and upload the pretrained CNN.
Step 6: Set network parameters, including the learning rate and weight initialization.
Step 7: Begin the training loop
for ξ = 1 to ε do
Randomly select a mini-batch from (size: β) from the Xtrain dataset.
Perform forward propagation and compute the loss E using the [removed]6)
loss function
| Figure 6:
| Figure 7:
Update the weights using the Adam optimizer, which combines the advantages of momentum and adaptive learning rates for faster convergence.
End
Step 8: After training, store the calculated weights in the database for future use or evaluation.
Step 9: Use the Xtest dataset to estimate the accuracy of the trained CNN models on the test data.
Step 10: Compute the final classification metrics such as accuracy, precision, recall, and F1-score to assess the model's performance on the test dataset.
Stop.
Results
All the model implementations are based on the open-source deep learning framework Keras. The experiments are conducted on a Ubuntu Linux server with a 3.40 GHz i7-3770 CPU (16 GB memory) and a GTX 1070 GPU (8 GB memory). The experiments are carried out with VGG-16, EfficientNet-B7, and CapsNet models. The models are optimized to make them compatible with the constructed image dataset and improve their performance. Separate training and testing have been conducted for each model.[11]
Performance Evaluation
The plot of classification efficiency of VGG-16, EfficientNet B-7, and CapsNet models based on cervical cancer stages is shown in [Fig. 3]. It is designed to help distinguish between tumors in stages 0 through 4 based on how far they have spread. When it comes to the classification of cervical cancer stages, the EfficientNet B-7 produces high accuracy compared with VGG-16 and CapsNet models. As illustrated in [Fig. 4], the EfficientNet B-7 outperforms VGG-16 and CapsNet models. According to the results of the testing, the EfficientNet B-7 is more accurate than VGG-16 and CapsNet models when it comes to categorizing different types of stages of cervical cancer.
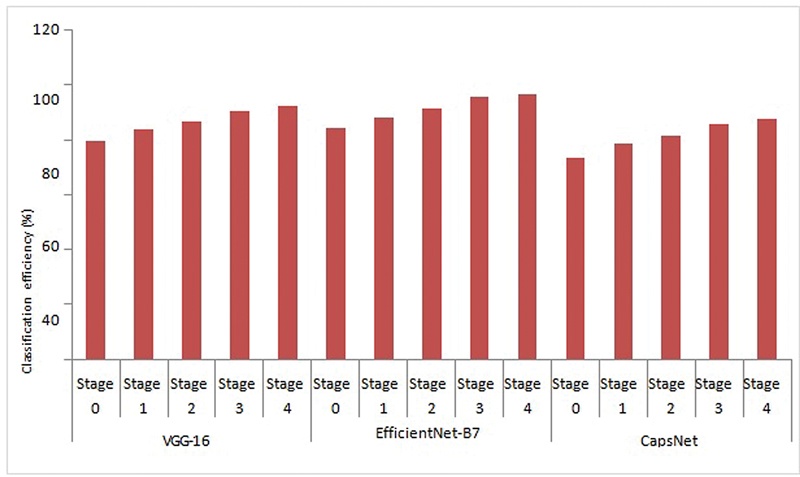
| Fig 3: Cervical cancer stage identification using CNN models.
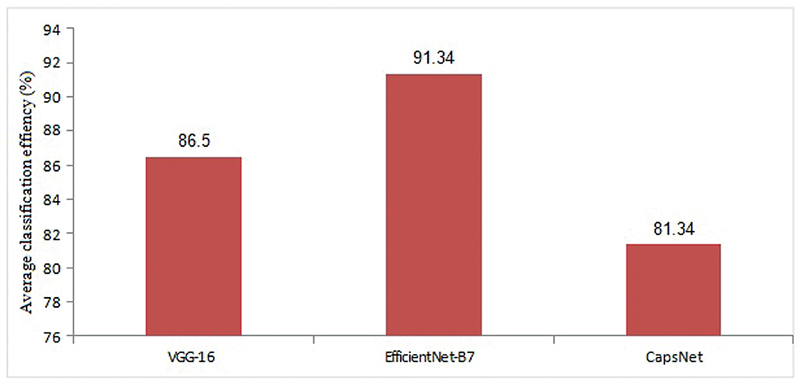
| fig 4: Average classification accuracy of cervical cancer stages using CNN models.
The performance metrics such as precision, recall, and F1-score are computed for the VGG-16, EfficientNet-B7, and CapsNet models and listed in [Supplementary Tables 4] to [6], (available in the online version only) respectively.
Discussion
As illustrated in [Fig. 3], the highest accuracy of 96.8% for stage 4 and lowest accuracy of 84.5% for stage 0 is achieved using Efficent B-7 model, the highest accuracy of 92.30% for stage 4 and lowest accuracy of 79.40% for stage 0 is achieved using VGG-16 model, and the highest accuracy of 87.40% for stage 4 and lowest accuracy of 73.50% for stage 0 is achieved using CapsNet model. From [Fig. 4], the Efficent B-7 model produces average classification efficiency of 91.34%, the VGG-16 model produces average classification efficiency of 86.5%, and the CapsNet model produces average classification efficiency of 81.34%. From the comparison results of CNN models, Efficient B-7 is superior for the classification of cervical cancer images considered in the present work.
As illustrated in [Supplementary Table 4], (available in the online version only) the precision of the EfficientNet B-7 compares favorably with the precision of VGG-16 and CapsNet CNN models. With repeated training on numerous images, precision can be reached with fewer errors. According to the results of the testing, EfficientNet B-7 beats the other models in terms of precision and is therefore recommended. The precision must be greater in order for the categorization to be correct; this means that there must be more precision in the classification result. [Supplementary Table 5] (available in the online version only) compares the sensitivity (recall) of the EfficientNet B-7 model to that of VGG-16 and CapsNet. As depicted, EfficientNet B-7 allows for the retrieval of a greater number of true positive cases than in the other cases. According to the findings of the testing, the EfficientNet B-7 model is more sensitive than the other models tested in this study. [Supplementary Table 6] (available in the online version only) gives a comparison of the F-measure of the EfficientNet B-7 with the F-measure of VGG-16 and CapsNet models. As depicted, EfficientNet B-7 outperforms VGG-16 and CapsNet models in terms of F-measure compared to the other models.
As demonstrated from the experimental results, the error rate of the EfficientNet B-7 is significantly much less than that of VGG-16 and CapsNet models. The EfficientNet B-7 is more accurate in categorizing cervical cancer stages than VGG-16 and CapsNet models. According to the results of the testing, the EfficientNet B-7 has a lower miss classification rate than the VGG-16 and CapsNet models.
Conclusion
This study presents a deep learning based approach for the categorization of cervical cancer stages using the SIPaKMeD dataset. By leveraging transfer learning with pretrained VGG-16, EfficientNet-B7, and CapsNet models, deep learning is demonstrated to significantly enhance the accuracy of cytological image classification. The dataset is expanded through classical augmentation techniques increasing its size to 24,294 images, thereby improving the generalization of the models. Among the models tested, EfficientNet-B7 achieved the highest classification accuracy outperforming VGG-16 and CapsNet CNN models in both training and validation phases. The use of ImageNet pretrained weights facilitated faster convergence and improved feature extraction, making the model well-suited for cervical cancer classification. The evaluation metrics such as precision, sensitivity, and F1-score confirmed the reliability of EfficientNet-B7 in distinguishing between different cancer stages. This work highlights the potential of deep learning in automating cervical cancer screening, reducing dependency on manual analysis, and improving early detection. Future work will focus on enhancing model interpretability, integrating additional datasets, exploring hybrid models, and optimizing computational efficiency for real-time clinical applications.
Conflict of Interest
None declared.
Acknowledgment
The authors thank Dr. Vishwas Pai, Consulting Oncologist, Pai Onco Care Centre, Hubballi, India, for his valuable suggestions.
Data Availability Statement
Datasets associated with this article are available at (https://universe.roboflow.com/ik-zu-quan-o9tdm/sipakmed-ioflq).
Patient's Consent
Patient consent was waived as the data used were anonymized and obtained from publicly available sources.
Supplementary Material
References
Address for correspondence
Publication History
Article published online:
03 July 2025
© 2025. The Author(s). This is an open access article published by Thieme under the terms of the Creative Commons Attribution License, permitting unrestricted use, distribution, and reproduction so long as the original work is properly cited. (https://creativecommons.org/licenses/by/4.0/)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
- Deep learning radiomics of multimodal ultrasound for classifying metastatic cervical lymphadenopathy into primary cancer sites: a feasibility studyYangyang Zhu, Ultraschall in der Medizin - European Journal of Ultrasound
- Artificial Intelligence for early identification of Amyloid positivity in transgenic Alzheimer miceF. Eckenweber, Nuklearmedizin, 2022
- Deep-learning-based clinical decision support system for gastric neoplasms in real-time endoscopy: Development and validation studyEun Jeong Gong, Endoscopy
- Advancing Medical Imaging Informatics by Deep Learning-Based Domain AdaptationAnirudh Choudhary, Yearbook of Medical Informatics
- Automated classification of gastric neoplasms in endoscopic images using a convolutional neural networkBum-Joo Cho, Endoscopy, 2019
- Graph-based methods for cervical cancer segmentation: Advancements, limitations, and future directions<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- Assessments of Data-Driven Deep Learning Models on One-Month Predictions of Pan-Arctic Sea Ice Thickness<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- Pancancer outcome prediction via a unified weakly supervised deep learning model<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- A data-driven method for microgrid bidding optimization in electricity market<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- Identification and comparison of organic matter-hosted pores in shale by SEM image analysis—a deep learning-based approach<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">

| Fig 1 : Schematic overview of the proposed method.

| fig 2:Stages of cervical cancer.
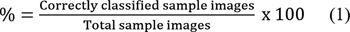
| Fig 1 :
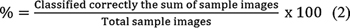
| Figure 2:
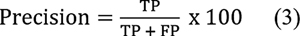
| Figure 3:
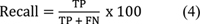
| Figure 4:
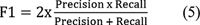
| Figure 5: (A)
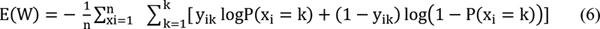
| Figure 6: (A)
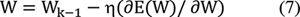
| Figure 7: (A)

| Fig 3: Cervical cancer stage identification using CNN models.

| fig 4: Average classification accuracy of cervical cancer stages using CNN models.


 PDF
PDF  Views
Views  Share
Share

