DROSHA rs642321 Polymorphism Influence Susceptibility to Childhood Acute Lymphoblastic Leukemia: A Preliminary Report
CC BY-NC-ND 4.0 · Indian J Med Paediatr Oncol 2017; 38(04): 416-419`
DOI: DOI: 10.4103/ijmpo.ijmpo_4_15
Abstract
Introduction: It has been well known that the microRNA biogenesis is involved in the pathogenesis of various diseases. We investigated the possible association between DROSHA rs642321 variant and risk of acute lymphocytic leukemia (ALL). Materials and Methods: We genotyped 75 children diagnosed with ALL and 115 age- and sex-matched children with no history of cancer of any type (as the control group) by the tetra amplification refractory mutation system-polymerase chain reaction. Results: We found that DROSHA rs642321 C > T variant significantly decreased the risk of ALL in codominant (TT vs. CC: odds ratio [OR] = 0.33, 95% confidence interval [CI] = 0.14–0.80, P = 0.020) and dominant (TT + CT vs. CC: OR = 0.51, 95% CI = 0.27–0.94, P = 0.037) inheritance model tested. The rs642321 T allele was associated with protective against ALL (OR = 0.58, 95% CI = 0.38–0.88, P = 0.011) incomparison with C allele. Conclusion: The study findings revealed that DROSHA rs642321 variant decreased the risk of pediatrics ALL in an Iranian population. Larger sample sizes with different ethnicities are needed to validate our findings.
Publication History
Article published online:
04 July 2021
© 2017. Indian Society of Medical and Paediatric Oncology. This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial-License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used forcommercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/.)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
Abstract
Introduction:
It has been well known that the microRNA biogenesis is involved in the pathogenesis of various diseases. We investigated the possible association between DROSHA rs642321 variant and risk of acute lymphocytic leukemia (ALL).
Materials and Methods:
We genotyped 75 children diagnosed with ALL and 115 age- and sex-matched children with no history of cancer of any type (as the control group) by the tetra amplification refractory mutation system-polymerase chain reaction.
Results:
We found that DROSHA rs642321 C > T variant significantly decreased the risk of ALL in codominant (TT vs. CC: odds ratio [OR] = 0.33, 95% confidence interval [CI] = 0.14–0.80, P = 0.020) and dominant (TT + CT vs. CC: OR = 0.51, 95% CI = 0.27–0.94, P = 0.037) inheritance model tested. The rs642321 T allele was associated with protective against ALL (OR = 0.58, 95% CI = 0.38–0.88, P = 0.011) in comparison with C allele.
Conclusion:
The study findings revealed that DROSHA rs642321 variant decreased the risk of pediatrics ALL in an Iranian population. Larger sample sizes with different ethnicities are needed to validate our findings.
Introduction
Acute lymphocytic leukemia (ALL) is the most common malignancy in childhood, and it is the main cause of morbidity and mortality in childhood. Although the pathogenesis of the disease is not fully understood, it is known that genetic polymorphisms play a critical role in ALL development.[1,2]
MicroRNAs (miRNAs) are a class of single-stranded, small noncoding RNA molecules, which are approximately 22 nucleotides in length.[3] They are involved in regulation of gene expression through binding to the 3'untranslated region (3'UTR) of target mRNAs, resulting in mRNA degradation or translational inhibition. It is now clear that miRNAs contribute to most diverse biological processes such as differentiation, cell proliferation, and apoptosis.[4,5] Besides, miRNAs can function as either oncogenes or tumor suppressors and play an important role in the pathogenesis of cancer.[6,7]
Biosynthesis of miRNAs involves various miRNA machinery genes and occurs in multiple steps. RNA polymerase II produces primary miRNA (~500–3000 nucleotides), which is processed by a multiprotein complex that includes DROSHA to form a precursor miRNA hairpin (~60–100 nucleotides, pre-miRNA). Then, pre-miRNA transported to the cytoplasm by Ran GTPase which subsequently processed by DICER1 to generate a single strand mature miRNA.[8,9,10,11]
The roles of miRNAs and miRNA biosynthesis pathway genes have been reported in cancers.[12,13,14,15,16,17] Several studies have shown the association between genetic polymorphisms in the miRNAs biogenesis genes and risk of various cancers.[15,18,19,20,21,22]
In this study, for the first time, we evaluated the impact of DROSHA rs642321 polymorphism on risk of childhood ALL in a sample of Iranian population.
Materials and Methods
Patients
In this case–control study, we recruit 75 children diagnosed with ALL and 115 age- and sex-matched healthy children in a southeast Iranian population. The study design and the enrolment procedure have been described previously.[23,24] Ethical approvals for recruitment were obtained from Local Ethics Committee of Zahedan University of Medical Sciences, and informed consent was obtained from parents of patients and healthy individuals in accordance with our institutional guidelines. Peripheral blood sample was collected in ethylenediamine tetraacetic acid-containing tubes from patients and healthy controls and DNA were extracted using salting out method as described previously.[25]
Genotyping
We used tetra amplification refractory mutation system – polymerase chain reaction (PCR), which is a simple and rapid method for detection of single nucleotide polymorphisms (SNPs),[26,27,28,29] for genotyping DROSHA rs642321 polymorphism.
We used four primers, two outer primers (forward outer: 5′-AAAGCTATAAACCCTCCTCCTTCCTGC-3′, reverse outer: 5′-CCCAGATTGTGACCCTAGACTTTCAATT-3′) and two allele-specific internal primers (forward inner [T allele]: 5′-CCCCTTCTCCCATTAGCCAATTACTT-3′, reverse inner [C allele]: 5′-GTGTTTTGTAGAAGGAATCCGTTTACCG-3′).
In each 0.20 ml PCR reaction tube, 1 μl of genomic DNA (~100 ng/ml), 1 μl of each primer (10 μM), 10 μl of 2 × Prime Taq Premix (Genet Bio, Korea) and 5 μl ddH2O were added.
Amplification was done with an initial denaturation step at 95°C for 5 min, followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C with a final step at 72°C for 10 min. PCR products were verified on a 2.5% agarose gel contained 0.5 μg/ml ethidium bromide, and photographs were taken [Figure 1]. Product sizes were 245 bp for T allele, 294 bp for C allele, and 486 bp for the two outer primers (control band). To verify genotyping quality, all polymorphisms in random samples were re-genotyped.
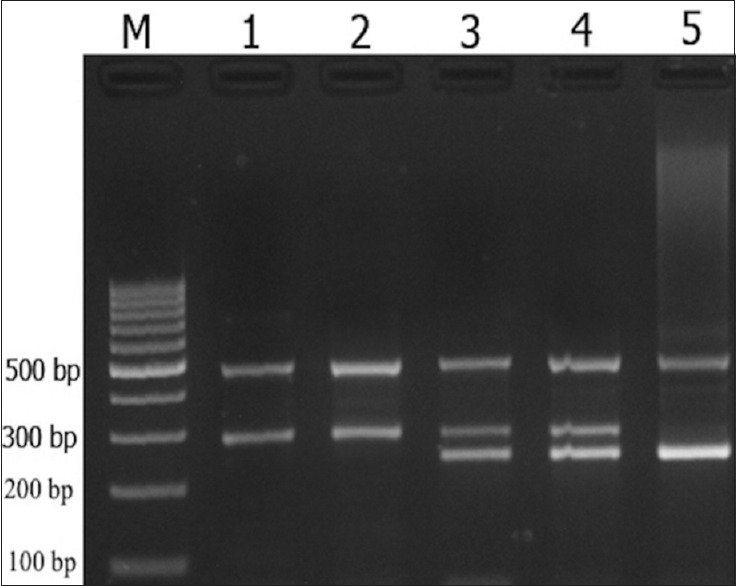
| Figure 1:Agarose gel electrophoresis pattern of tetra amplification refractory mutation system polymerase chain reaction (T-ARMS-PCR) for detection of DROSHA rs642321 polymorphism. Product sizes were 245 bp for T allele, 294 bp for C allele, and 486 bp for the two outer primers. M: DNA marker; Lanes 1,2: rs642321 CC; Lanes 3,4: rs642321 CT; Lane 5: rs642321 TT
Statistical analysis
Statistical analyses were performed using the SPSS software for Windows, version 18.0 (SPSS Inc., Chicago IL, USA). Student's t-test and ANOVA was used to test for differences in continuous variables. The categorical data were analyzed using χ2 test. The values of P < 0 xss=removed>2 test in both cases and controls.
Results
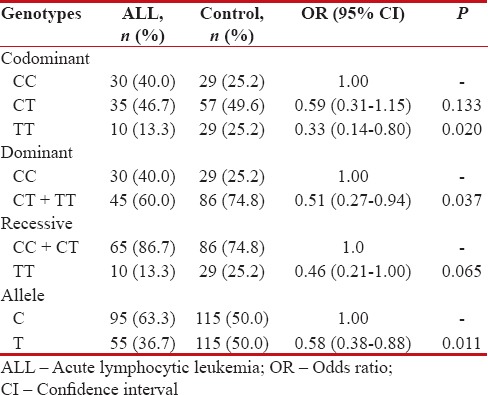
In this study, we genotyped DROSHA rs642321 polymorphism in 75 ALL children and 115 healthy children. The findings demonstrated that DROSHA rs642321 C > T variant significantly decreased the risk of ALL in codominant (TT vs. CC: odds ratio [OR] = 0.33, 95% CI = 0.14–0.80, P =0.020) and dominant (TT + CT vs. CC: OR = 0.51, 95% CI = 0.27–0.94, P =0.037) inheritance model tested. The rs642321 T allele was associated with protective against ALL (OR = 0.58, 95% CI = 0.38–0.88, P = 0.011) in comparison with C allele [Table 1]. The DROSHA rs642321 genotypes in cases and controls were in HWE (χ2 = 0.002, P = 0.966, and χ2 = 0.008, P = 0.925, respectively).
Table 1
Genotypic and allelic frequency of DROSHA rs642321 polymorphism in childhood acute lymphocytic leukemia and control subjects
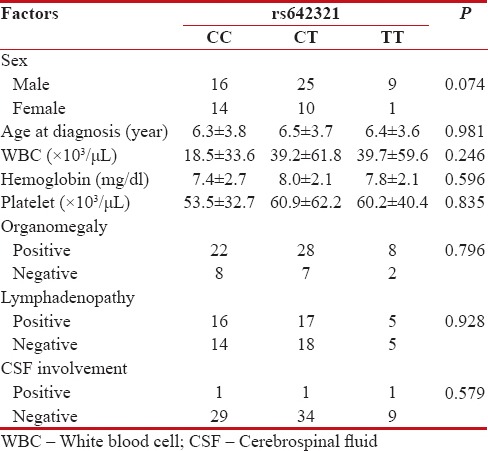
| We analyzed possible association between DROSHA rs642321 variant and patients' demographic and clinical data. As shown in Table 2, no association was found between ages of patients, white blood cell count, hemoglobin concentration, platelet count, sex, organomegaly, lymphadenopathy as well as cerebrospinal fluid involvement and rs642321 genotypes.
Table 2
Association of DROSHA rs642321gene polymorphism with demographic and clinical features of childhood acute lymphocytic leukemia patients
|
Discussion
In recent years, it has been revealed that miRNAs are important components of complex gene regulatory networks. miRNA are a class of small noncoding RNA molecules involved in a variety of cellular functions.[5] SNPs in miRNA sequences, miRNA target genes as well as miRNA biogenesis may affect the miRNAs regulation.[17,18,30,31]
In this study, we aimed to find out the impact of DROSHA rs642321 polymorphism on childhood ALL. The study findings showed an association between DROSHA rs642321 and ALL. The DROSHA rs642321 TT as well as TT + CT genotype decreased the risk of ALL in comparison with CC genotype. The rs642321 T allele was significantly associated with protective against ALL. Gutierrez-Caminors et al.[31] have found that DROSHA rs10035440 variant increased the risk of pediatric ALL.
DROSHA is a member of RNase III superfamily and playing a pivotal role in the pathway of miRNAs biogenesis.[32,33] Sung et al.[20] evaluated the impact of rs644236, rs874332, rs7737174, rs10461898, rs11748548, and rs16901096 polymorphisms of DROSHA on breast cancer and found no significant association between these variants and breast cancer risk. They reported that DROSHA rs644236 TT genotype as well as rs7737174 AA genotype was associated with breast cancer risk in postmenopausal women.[20] Weng et al.[21] have found an association between DROSHA rs10719 variant and risk of malignant peripheral nerve sheath tumor.
Lin et al.[34] stated the haplotypes of DROSHA and DICER, but not individual variant, is associated with altered survival and recurrence of renal cell carcinoma. Yuan et al.[18] have found no significant association between DROSHA rs642321 polymorphism and bladder cancer risk, while rs10719T > C variant increased the risk of the disease in a Chinese population.
The aberrant expression of DROSHA at mRNA or protein level has been reported in numerous types of cancer, including skin cancer,[35] cervical cancer,[36] ovarian cancer,[15] breast cancer,[37] nonsmall-cell lung cancer,[38] and neuroblastoma.[39]
Conclusion
The study finding showed that rs642321 variant located in DROSHA 3'UTR decreased the risk of childhood ALL. The association and functional study are necessary to certify our result.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
This paper was based on MSc thesis of SSH (#5696), and the deputy for Research at Zahedan University of Medical Sciences provided the fund.
References
- Guo LM, Xi JS, Ma Y, Shao L, Nie CL, Wang GJ, et al. ARID5B gene rs10821936 polymorphism is associated with childhood acute lymphoblastic leukemia: A meta-analysis based on 39,116 subjects. Tumour Biol 2014;35:709-13.
- Ma Y, Sui Y, Wang L, Li H. Effect of GSTM1 null genotype on risk of childhood acute leukemia: A meta-analysis. Tumour Biol 2014;35:397-402.
- Ambros V. MicroRNAs: Tiny regulators with great potential. Cell 2001;107:823-6.
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet 2008;9:102-14.
- Wilczynska A, Bushell M. Thecomplexity of miRNA-mediated repression. Cell Death Differ 2015;22:22-33.
- Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302:1-2.
- Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 2005;353:1768-71.
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5.
- Bartel DP, Chen CZ. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004;5:396-400.
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science 2004;303:95-8.
- ;Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science 2005;309:1519-24.
- Yan M, Huang HY, Wang T, Wan Y, Cui SD, Liu ZZ, et al. Dysregulated expression of dicer and drosha in breast cancer. Pathol Oncol Res 2012;18:343-8.
- Sung H, Jeon S, Lee KM, Han S, Song M, Choi JY, et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and breast cancer survival. BMC Cancer 2012;12:195.
- Jafarnejad SM, Sjoestroem C, Martinka M, Li G. Expression of the RNase III enzyme DROSHA is reduced during progression of human cutaneous melanoma. Mod Pathol 2013;26:902-10.
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, drosha, and outcomes in patients with ovarian cancer. N Engl J Med 2008;359:2641-50.
- Zhu DX, Fan L, Lu RN, Fang C, Shen WY, Zou ZJ, et al. Downregulated dicer expression predicts poor prognosis in chronic lymphocytic leukemia. Cancer Sci 2012;103:875-81.
- Tong N, Xu B, Shi D, Du M, Li X, Sheng X, et al. Hsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese children. Mutat Res 2014;759:16-21.
- Yuan L, Chu H, Wang M, Gu X, Shi D, Ma L, et al. Genetic variation in DROSHA 3'UTR regulated by hsa-miR-27b is associated with bladder cancer risk. PLoS One 2013;8:e81524.
- Jiang Y, Chen J, Wu J, Hu Z, Qin Z, Liu X, et al. Evaluation of genetic variants in microRNA biosynthesis genes and risk of breast cancer in chinese women. Int J Cancer 2013;133:2216-24.
- Sung H, Lee KM, Choi JY, Han S, Lee JY, Li L, et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and risk of breast cancer: A case-control study in Korea. Breast Cancer Res Treat 2011;130:939-51.
- Weng Y, Chen Y, Chen J, Liu Y, Bao T. Common genetic variants in the microRNA biogenesis pathway are associated with malignant peripheral nerve sheath tumor risk in a Chinese population. Cancer Epidemiol 2013;37:913-6.
- Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res 2008;14:7956-62.
- Hashemi M, Sheybani-Nasab M, Naderi M, Roodbari F, Taheri M. Association of functional polymorphism at the miR-502-binding site in the 3' untranslated region of the SETD8 gene with risk of childhood acute lymphoblastic leukemia, a preliminary report. Tumour Biol 2014;35:10375-9.
- Hasani SS, Hashemi M, Eskandari-Nasab E, Naderi M, Omrani M, Sheybani-Nasab M, et al. Afunctional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: A preliminary report. Tumour Biol 2014;35:219-25.
- Hashemi M, Moazeni-Roodi AK, Fazaeli A, Sandoughi M, Bardestani GR, Kordi-Tamandani DM, et al. Lack of association between paraoxonase-1 Q192R polymorphism and rheumatoid arthritis in Southeast Iran. Genet Mol Res 2010;9:333-9.
- Hashemi M, Fazaeli A, Ghavami S, Eskandari-Nasab E, Arbabi F, Mashhadi MA, et al. Functional polymorphisms of FAS and FASL gene and risk of breast cancer – Pilot study of 134 cases. PLoS One 2013;8:e53075.
- Hashemi M, Eskandari-Nasab E, Zakeri Z, Atabaki M, Bahari G, Jahantigh M, et al. Association of pre-miRNA-146a rs2910164 and premiRNA-499 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Mol Med Rep 2013;7:287-91.
- Hashemi M, Eskandari-Nasab E, Moazeni-Roodi A, Naderi M, Sharifi-Mood B, Taheri M, et al. Association of CTSZ rs34069356 and MC3R rs6127698 gene polymorphisms with pulmonary tuberculosis. Int J Tuberc Lung Dis 2013;17:1224-8.
- Hashemi M, Moazeni-Roodi A, Bahari A, Taheri M. A tetra-primer amplification refractory mutation system-polymerase chain reaction for the detection of rs8099917 IL28B genotype. Nucleosides Nucleotides Nucleic Acids 2012;31:55-60.
- Kan CW, Howell VM, Hahn MA, Marsh DJ. Genomic alterations as mediators of miRNA dysregulation in ovarian cancer. Genes Chromosomes Cancer 2015;54:1-9.
- Gutierrez-Camino A, Lopez-Lopez E, Martin-Guerrero I, Piñan MA, Garcia-Miguel P, Sanchez-Toledo J, et al. Noncoding RNA-related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr Res 2014;75:767-73.
- Mueller GA, Miller MT, DeRose EF, Ghosh M, London RE, Hall TM, et al. Solution structure of the drosha double-stranded RNA-binding domain. Silence 2010;1:2.
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III drosha initiates microRNA processing. Nature 2003;425:415-9.
- Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X, et al. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis 2010;31:1805-12.
- Sand M, Gambichler T, Skrygan M, Sand D, Scola N, Altmeyer P, et al. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Invest 2010;28:649-53.
- Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, et al. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol 2007;212:368-77.
- Avery-Kiejda KA, Braye SG, Forbes JF, Scott RJ. The expression of dicer and Drosha in matched normal tissues, tumours and lymph node metastases in triple negative breast cancer. BMC Cancer 2014;14:253.
- Lønvik K, Sørbye SW, Nilsen MN, Paulssen RH. Prognostic value of the microRNA regulators Dicer and Drosha in non-small-cell lung cancer: Co-expression of drosha and miR-126 predicts poor survival. BMC Clin Pathol 2014;14:45.
- Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, et al. MicroRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res 2010;70:7841-50.

| Figure 1:Agarose gel electrophoresis pattern of tetra amplification refractory mutation system polymerase chain reaction (T-ARMS-PCR) for detection of DROSHA rs642321 polymorphism. Product sizes were 245 bp for T allele, 294 bp for C allele, and 486 bp for the two outer primers. M: DNA marker; Lanes 1,2: rs642321 CC; Lanes 3,4: rs642321 CT; Lane 5: rs642321 TT
References
- Guo LM, Xi JS, Ma Y, Shao L, Nie CL, Wang GJ, et al. ARID5B gene rs10821936 polymorphism is associated with childhood acute lymphoblastic leukemia: A meta-analysis based on 39,116 subjects. Tumour Biol 2014;35:709-13.
- Ma Y, Sui Y, Wang L, Li H. Effect of GSTM1 null genotype on risk of childhood acute leukemia: A meta-analysis. Tumour Biol 2014;35:397-402.
- Ambros V. MicroRNAs: Tiny regulators with great potential. Cell 2001;107:823-6.
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet 2008;9:102-14.
- Wilczynska A, Bushell M. Thecomplexity of miRNA-mediated repression. Cell Death Differ 2015;22:22-33.
- Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302:1-2.
- Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 2005;353:1768-71.
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5.
- Bartel DP, Chen CZ. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004;5:396-400.
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science 2004;303:95-8.
- ;Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science 2005;309:1519-24.
- Yan M, Huang HY, Wang T, Wan Y, Cui SD, Liu ZZ, et al. Dysregulated expression of dicer and drosha in breast cancer. Pathol Oncol Res 2012;18:343-8.
- Sung H, Jeon S, Lee KM, Han S, Song M, Choi JY, et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and breast cancer survival. BMC Cancer 2012;12:195.
- Jafarnejad SM, Sjoestroem C, Martinka M, Li G. Expression of the RNase III enzyme DROSHA is reduced during progression of human cutaneous melanoma. Mod Pathol 2013;26:902-10.
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, drosha, and outcomes in patients with ovarian cancer. N Engl J Med 2008;359:2641-50.
- Zhu DX, Fan L, Lu RN, Fang C, Shen WY, Zou ZJ, et al. Downregulated dicer expression predicts poor prognosis in chronic lymphocytic leukemia. Cancer Sci 2012;103:875-81.
- Tong N, Xu B, Shi D, Du M, Li X, Sheng X, et al. Hsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese children. Mutat Res 2014;759:16-21.
- Yuan L, Chu H, Wang M, Gu X, Shi D, Ma L, et al. Genetic variation in DROSHA 3'UTR regulated by hsa-miR-27b is associated with bladder cancer risk. PLoS One 2013;8:e81524.
- Jiang Y, Chen J, Wu J, Hu Z, Qin Z, Liu X, et al. Evaluation of genetic variants in microRNA biosynthesis genes and risk of breast cancer in chinese women. Int J Cancer 2013;133:2216-24.
- Sung H, Lee KM, Choi JY, Han S, Lee JY, Li L, et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and risk of breast cancer: A case-control study in Korea. Breast Cancer Res Treat 2011;130:939-51.
- Weng Y, Chen Y, Chen J, Liu Y, Bao T. Common genetic variants in the microRNA biogenesis pathway are associated with malignant peripheral nerve sheath tumor risk in a Chinese population. Cancer Epidemiol 2013;37:913-6.
- Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res 2008;14:7956-62.
- Hashemi M, Sheybani-Nasab M, Naderi M, Roodbari F, Taheri M. Association of functional polymorphism at the miR-502-binding site in the 3' untranslated region of the SETD8 gene with risk of childhood acute lymphoblastic leukemia, a preliminary report. Tumour Biol 2014;35:10375-9.
- Hasani SS, Hashemi M, Eskandari-Nasab E, Naderi M, Omrani M, Sheybani-Nasab M, et al. Afunctional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: A preliminary report. Tumour Biol 2014;35:219-25.
- Hashemi M, Moazeni-Roodi AK, Fazaeli A, Sandoughi M, Bardestani GR, Kordi-Tamandani DM, et al. Lack of association between paraoxonase-1 Q192R polymorphism and rheumatoid arthritis in Southeast Iran. Genet Mol Res 2010;9:333-9.
- Hashemi M, Fazaeli A, Ghavami S, Eskandari-Nasab E, Arbabi F, Mashhadi MA, et al. Functional polymorphisms of FAS and FASL gene and risk of breast cancer – Pilot study of 134 cases. PLoS One 2013;8:e53075.
- Hashemi M, Eskandari-Nasab E, Zakeri Z, Atabaki M, Bahari G, Jahantigh M, et al. Association of pre-miRNA-146a rs2910164 and premiRNA-499 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Mol Med Rep 2013;7:287-91.
- Hashemi M, Eskandari-Nasab E, Moazeni-Roodi A, Naderi M, Sharifi-Mood B, Taheri M, et al. Association of CTSZ rs34069356 and MC3R rs6127698 gene polymorphisms with pulmonary tuberculosis. Int J Tuberc Lung Dis 2013;17:1224-8.
- Hashemi M, Moazeni-Roodi A, Bahari A, Taheri M. A tetra-primer amplification refractory mutation system-polymerase chain reaction for the detection of rs8099917 IL28B genotype. Nucleosides Nucleotides Nucleic Acids 2012;31:55-60.
- Kan CW, Howell VM, Hahn MA, Marsh DJ. Genomic alterations as mediators of miRNA dysregulation in ovarian cancer. Genes Chromosomes Cancer 2015;54:1-9.
- Gutierrez-Camino A, Lopez-Lopez E, Martin-Guerrero I, Piñan MA, Garcia-Miguel P, Sanchez-Toledo J, et al. Noncoding RNA-related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr Res 2014;75:767-73.
- Mueller GA, Miller MT, DeRose EF, Ghosh M, London RE, Hall TM, et al. Solution structure of the drosha double-stranded RNA-binding domain. Silence 2010;1:2.
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III drosha initiates microRNA processing. Nature 2003;425:415-9.
- Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X, et al. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis 2010;31:1805-12.
- Sand M, Gambichler T, Skrygan M, Sand D, Scola N, Altmeyer P, et al. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Invest 2010;28:649-53.
- Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, et al. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol 2007;212:368-77.
- Avery-Kiejda KA, Braye SG, Forbes JF, Scott RJ. The expression of dicer and Drosha in matched normal tissues, tumours and lymph node metastases in triple negative breast cancer. BMC Cancer 2014;14:253.
- Lønvik K, Sørbye SW, Nilsen MN, Paulssen RH. Prognostic value of the microRNA regulators Dicer and Drosha in non-small-cell lung cancer: Co-expression of drosha and miR-126 predicts poor survival. BMC Clin Pathol 2014;14:45.
- Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, et al. MicroRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res 2010;70:7841-50.


 PDF
PDF  Views
Views  Share
Share

