Retrospective Study of B Lymphoblastic Leukemia to Assess the Prevalence of TEL/AML1 in South India: A Study of 214 Cases and Review of Literature
CC BY-NC-ND 4.0 · Indian J Med Paediatr Oncol 2025; 46(03): 259-268
DOI: DOI: 10.1055/s-0042-1742611
Abstract
Introduction Translocation t(12;21)(p13;q22), a recurrent and an invisible chromosomal abnormality, resulting in TEL/AML1 gene fusion, associated with good prognosis, has been described to be a common abnormality, in children with B-acute lymphoblastic leukemia (B-ALL).
Objectives The initial observation of very few TEL/AML1 positive patients at this center on testing by fluorescence in situ hybridization (FISH) led to study the prevalence of the abnormality, compare with the global distribution, and evaluate clinical, pathological, molecular, and cytogenetic features in TEL/AML1 positive patients.
Materials and Methods A retrospective study of all B-ALL patients tested for TEL/AML1 gene fusion during the period January 2009 to November 2020 was undertaken. Clinicopathological, molecular, cytogenetic, treatment, and follow-up details were collected. All publications dealing with TEL/AML1 gene rearrangement were reviewed post Google and PubMed search.
Results TEL/AML1gene rearrangement was assessed by FISH in 178 patients and by reverse transcription polymerase chain reaction in 36 patients and detected as the sole abnormality in 8.4% patients with additional genetic abnormalities noted on FISH evaluation. Normal karyotype was noted in 14/18 (77.7%) of these patients and 2 had complex karyotype. Complete blood count revealed hemoglobin to range from 35 to 116 g/L (median: 74 g/L), white blood count: 1.01–110×109/L (median: 7.8×109/L), platelet counts: 10–115×109/L (median: 42×109/L), blast count in peripheral smear: 0–98% (median: 41%). Immunophenotyping demonstrated 94.4% were CD34 positive, common acute lymphoblastic leukemia associated antigen (CALLA) positive with aberrant expression of CD13, CD33, CD56, singly or in combination in 58.8%.
Conclusion TEL/AML1 fusion is rare in Indian patients with B-ALL and appears to be much rarer in our region. The detection of relevant specific abnormalities is of fundamental importance in B-ALL patients and these geographic variations can be used in defining management policies.
Keywords
TEL/AML1 - t(12;21) fluorescence in situ hybridization - acute lymphoblastic leukemia (ALL) - RT-PCRPublication History
Article published online:
20 May 2022
© 2022. Indian Society of Medical and Paediatric Oncology. This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used for commercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/)
Thieme Medical and Scientific Publishers Pvt. Ltd.
A-12, 2nd Floor, Sector 2, Noida-201301 UP, India
- Aberrantly regulated genes in TEL/AML1 positive leukaemiaU. Hentschel, Klinische Pädiatrie, 2004
- Frequency of 11q23/MLL gene rearrangement in Egyptian childhood acute myeloblastic leukemia: Biologic and clinical significanceAdelAbdelhaleim Hagag, South Asian Journal of Cancer, 2014
- Cytogenetics and Molecular Genetics in Pediatric Acute Lymphoblastic Leukemia (ALL) and Its Correlation with Induction OutcomesAjeitha Loganathan, South Asian Journal of Cancer
- Rare and recurrent chromosomal abnormalities and their clinical relevance in pediatric acute leukemia of south Indian populationSeyed Hashem Mir Mazloumi, Indian Journal of Medical and Paediatric Oncology, 2012
- Characterization of the t(7;12) chromosomal translocation in infant AMLs: coexpression of T-cell markers, distinct expression of homeobox genes and inhibition o...S Wildenhain, Klinische Pädiatrie, 2009
- Pulmonary arteriovenous malformations: an uncharacterised phenotype of dyskeratosis congenita and related telomere biology disorders<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- First heterozygous NOP10 mutation in Familial Pulmonary Fibrosis<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- Size matters! Peripheral blood leukocyte telomere length and survival after critical illness<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- Pathology and radiology of interstitial lung disease in patients with telomere-related genes mutations<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
- Telomere length in preterm-born children with bronchopulmonary dysplasia (BPD)<svg viewBox="0 0 24 24" fill="none" xmlns="http://www.w3.org/2000/svg">
Abstract
Introduction Translocation t(12;21)(p13;q22), a recurrent and an invisible chromosomal abnormality, resulting in TEL/AML1 gene fusion, associated with good prognosis, has been described to be a common abnormality, in children with B-acute lymphoblastic leukemia (B-ALL).
Objectives The initial observation of very few TEL/AML1 positive patients at this center on testing by fluorescence in situ hybridization (FISH) led to study the prevalence of the abnormality, compare with the global distribution, and evaluate clinical, pathological, molecular, and cytogenetic features in TEL/AML1 positive patients.
Materials and Methods A retrospective study of all B-ALL patients tested for TEL/AML1 gene fusion during the period January 2009 to November 2020 was undertaken. Clinicopathological, molecular, cytogenetic, treatment, and follow-up details were collected. All publications dealing with TEL/AML1 gene rearrangement were reviewed post Google and PubMed search.
Results TEL/AML1gene rearrangement was assessed by FISH in 178 patients and by reverse transcription polymerase chain reaction in 36 patients and detected as the sole abnormality in 8.4% patients with additional genetic abnormalities noted on FISH evaluation. Normal karyotype was noted in 14/18 (77.7%) of these patients and 2 had complex karyotype. Complete blood count revealed hemoglobin to range from 35 to 116 g/L (median: 74 g/L), white blood count: 1.01–110×109/L (median: 7.8×109/L), platelet counts: 10–115×109/L (median: 42×109/L), blast count in peripheral smear: 0–98% (median: 41%). Immunophenotyping demonstrated 94.4% were CD34 positive, common acute lymphoblastic leukemia associated antigen (CALLA) positive with aberrant expression of CD13, CD33, CD56, singly or in combination in 58.8%.
Conclusion TEL/AML1 fusion is rare in Indian patients with B-ALL and appears to be much rarer in our region. The detection of relevant specific abnormalities is of fundamental importance in B-ALL patients and these geographic variations can be used in defining management policies.
Keywords
TEL/AML1 - t(12;21) fluorescence in situ hybridization - acute lymphoblastic leukemia (ALL) - RT-PCRIntroduction
Detection of recurrent genetic abnormalities in hematological malignancies by fluorescence in situ hybridization (FISH) is part of routine standard of care. Recurrent cryptic translocation t(12;21)(p13;q22) in pediatric B-acute lymphoblastic leukemia (B-ALL) is associated with specific clinicopathological characteristics and favorable prognosis.[1] [2] [3] [4] Assessment for TEL/AML1(ETV6/RUNX1) fusion enables proper treatment strategies in a given population. Review of online hospital-based reports, international projects, multicenter studies, and review articles[3] [4] [5] demonstrated a wide variation in global incidence, with incidence in west being ∼25%[4] and lesser in India ([Table S1] and [S2-supplementary material], available in the online version).[5] [6] [7] [8] [9] [10] [11] [12] [13] [14] [15] [16] [17] [18] [19] [20] [21] [22] [23] [24] [25] [26] [27] [28] [29] [30] However, the initial observation of very less number of prognostically favorable TEL/AML1 rearranged B-ALL patients from our tertiary care cancer center in south India prompted focus on evaluating the prevalence of this abnormality. The aim and objectives of the study were to see the prevalence of TEL/AML1 gene fusion in newly diagnosed B-ALL patients, compare with the global distribution of the fusion gene, and evaluate the clinical, pathological, molecular, and cytogenetic features where available in TEL/AML1 positive patients.
Materials and Methods
A retrospective study of all cases, previously diagnosed as B-ALL during the period January 2009 to November 2020, at a tertiary care cancer center from South India, was undertaken. Data on demographic, clinical, hematological, interphase FISH, cytogenetics, reverse transcription polymerase chain reaction (RT-PCR) findings, treatment protocol, and follow-up details were collected from the case files in a format. All the newly diagnosed cases for which ALL prognostic panel (BCR-ABL, TEL-AML1, MLL) was performed and TEL AML result was available were included in the study. All cases where adequate blasts counts were not evident and also relapsed ALL cases were excluded from the study. Primary outcome provides information on the prevalence of TEL/AML1 gene fusion positivity in patients diagnosed with B-ALL from this region and for understanding how the prevalence compares with the global incidence for managing the patients. Secondary outcome includes defining the clinicopathlogic features, cytogenetics and FISH findings, baseline lab values and overall survival in the TEL AML1 positive patients that were analyzed.
FISH: Interphase FISH studies were performed till November 2019, on heparinized bone marrow/peripheral blood fixed samples in 178 patients. LSI ETV6(TEL)/RUNX1(AML1) extra signal dual color translocation probe set and ZytoLightETV6/RUNX1(TEL/AML1) dual color dual fusion probe were used for testing in 81and 97 samples, respectively. The probe details, typical signal patterns and interpretation are provided in [Table S3] ([Supplementary material], available in the online version). The denaturation and hybridization procedures were performed according to the manufacturer's instructions and counterstained with 4,6-diamino-2-phenyl-indole stain. Fluorescent signals were visualized at 1250 × magnification. The cutoff for TEL/AML1 positivity was 5%. For each case, 200 interphase cells were evaluated by two analysts.
The status of BCR ABL fusion and MLL gene rearrangement was also evaluated using appropriate probes ([Table S3], available in the online version).
RT-PCR: TEL/AML1 mutation was assessed by RT-PCR in 36 patient samples from December 2019 onward. RNA was extracted from bone marrow/peripheral blood samples in ethylenediaminetetraacetic acid, using Qiagen RNA blood mini kit and, c-DNA synthesis and real time PCR was performed using TRU-PCR ALL Panel Kit Version 2.1 (3B Black Biotech India Ltd, Bhopal, India).
Data on karyotype was available in 92 ALL cases. Twenty-four or forty-eight-hour unstimulated heparinized bone marrow/peripheral blood cultures had been set up as per standard protocol. Twenty metaphases were evaluated, and karyotype reported as per International System for Human Cytogenomic Nomenclature (2016).[31] Correlation with cytogenetics was done where available.
Statistical Analysis
Since the number of TEL/AML1 positive cases were few, simple statistical measure, percentage was used to express the prevalence and other data.
Ethical Approval
The procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation and with the Helsinki Declaration of 1964, as revised in 2013. Patient consent was taken prior to the enrolment of participants in the study. Approval of the study was obtained from Basavatarakam Indo American cancer Hospital & Research Institute's ethical committee. (EC Reference study code: 96, dated 27 May 2021).
Results
The patient cohort consisted of 214 B-ALL patients, at diagnosis, both adults and children. Data on demographic, clinical hematological, FISH, and karyotype findings are presented in [Tables 1], [2], and [3]. A total of 8.4% (18/214) patients harbored the TEL/AML1 gene rearrangement. The fusion was detected by FISH in 15/178(8.4%) patients and by RT-PCR in 3/36 (8.3%) patients. The clone size or positivity rate varied from 8.9 to 95%. The fusion was frequent in 17/125 (13.6%) children, aged between 2 and 13 years (median: 4 years), and an 18-year-old young adult. Fusion was noted in 94.4% males ([Table 1]).
|
Case no |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
17 |
18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Age(y) and gender M/F |
5 M |
3 M |
4 M |
4 M |
2 M |
4 M |
4 F |
18 M |
5 M |
2 M |
8 M |
4 M |
13 M |
5 M |
3 M |
8 M |
6 M |
7 M |
|
Clinical presentation and history |
Fever and weakness—2 months |
Fever and weakness—1 week |
Fever—2 weeks and multiple petechial rashes |
Fever—1 week and multiple joint pains |
Fever—2 weeks and joint pain for 1 week |
Fever—1 month, bilateral cervical LN, S+. |
Fever—1 month, cough cold—vomiting petechial rash—1 day.H+S+ |
Fever and AD |
Fever and weakness—1 week. |
Fever—2 weeks and multiple petechial rashes |
Fever—1 month, cough cold, vomiting, petechial rash—1 day. H+S+ |
Fever—1 week and multiple joint pains |
Fever—2 weeks, joint pain—1 week, bilateral cervical LN, S+ |
Fever—1 month bilateral cervical LN,S+ |
Fever and pallor |
Fever—1 week vomitting |
Fever—2weeks Cough cold |
Fever >1 month |
|
HB g/L: |
73 |
62 |
89 |
35 |
69 |
48 |
97 |
85 |
74 |
91 |
116 |
89 |
77 |
74 |
69 |
88 |
76 |
7 |
|
WBCX10(12)/L: |
9.7 |
110 |
3.2 |
36.7 |
5.92 |
4.5 |
6.8 |
17.95 |
4.59 |
1.91 |
6.12 |
3.66 |
8.9 |
1.01 |
50.31 |
17.5 |
11.22 |
23.62 |
|
Platelet x10(9)/L: |
100 |
10 |
45 |
12 |
86 |
17 |
65 |
89 |
22 |
74 |
115 |
24 |
15 |
62 |
18 |
50 |
39 |
45 |
|
PS smear blasts (%) |
25 |
95 |
10 |
90 |
18 |
6 |
50 |
46 |
0 |
30 |
36 |
65 |
4 |
98 |
70 |
56 |
80 |
|
|
BM blasts (%) |
85 |
90 |
77 |
88 |
95 |
60 |
95 |
58 |
80 |
88 |
40 |
69 |
77 |
3 |
55 |
|||
|
PAS |
+ |
+ |
NC |
ND |
+ |
+ |
+ |
N |
G + |
|||||||||
|
B-ALL CD10 +/− |
+ |
+ |
+ |
+ |
− |
+ |
+ |
+ |
+ |
|||||||||
|
Aberrant markers |
CD13 & CD33 |
CD56 |
CD13 & CD33 |
CD13 & CD33 |
CD13 |
CD13 |
||||||||||||
|
TEL AML1 |
*P |
*P |
P |
P |
P |
P |
P |
P |
P |
P |
P |
P |
P |
P |
P |
P# |
P# |
P# |
|
Signal pattern |
AT |
AT |
AT |
AT |
AT |
AT |
AT |
AT |
AT |
T |
AT |
AT |
AT |
AT |
AT |
|||
|
FISH signal pattern |
1G 2O3F |
2O1F |
1G1O1F |
3G1O1F |
2G2O1F. 2G 1O 1F |
2G1O1F |
2G1O1F |
2G2O2F |
1G1O1F, |
1G1O2F, |
2G2O2F, |
2G1O1F, |
1G1O1F, |
1G1O1F |
2G2O1F, 3G3O |
|||
|
Interpretation |
1 |
2 |
3 |
3 + 4(1-2) |
6(1)−3+ 4(1) +5(1) 6(2)−3+ 4(1) |
3 + 4 (1) |
3+ 4 (1) |
4(1) + 5(1) |
3 |
4(1) + 5(1) |
3 + 4(1) |
3 |
3 |
6(1)- P 3+ 4(2) +5(1) 6(2)- N, 4(1) +5(1) |
||||
|
Clone no and size (%) |
39 |
61 |
10 |
89 |
38 20 |
47 |
42 |
83 |
95 |
64 |
91 |
11 |
8.9 |
5 |
P 16 N |
|||
|
BCR-ABL, MLL, E2A-PBX |
N N |
N N |
N N |
N N |
ND,N |
ND, N |
N, N, N |
ND, N |
ND, N loss of MLL −91% |
ND,N |
ND,N |
#N , N |
ND, MLL -ve |
# N, N |
ND,N |
ND |
ND |
ND |
|
Karyotype |
NK |
NK |
NK |
NK |
IM |
NK |
NK |
NK |
NK |
NK |
NK |
* 46 |
NK |
NK |
NK |
AK |
NM |
AK |
Abbreviations: #, TEL AML1/BCR-ABL(p210,p190)-RT-PCR; +/G + , block/granular positive; AD, abdominal distension; B-ALL, B-acute lymphoblastic leukemia; FISH, fluorescence in situ hybridization; H+S + , hepatosplenomegaly; LN, lymphadenopathy; N, negative; NC, noncontributory; ND, not done; P, positive; RT-PCR, reverse transcription polymerase chain reaction; S + , splenomegaly
*Cases 1 & 2 (Vysis Probe) &, 3–15 (Zytovision Probe). Signal: green: G, orange: O, fusion: F, typical signal pattern:(T), normal nuclei—2G2O, abnormal nuclei with fusion—1G2O1F (Vysis)/ IGIO2F (Zytovision). Atypical signal pattern: AT, Interpretation: 1—extra fusion genes TEL/ AML1 on der 21q, 2—Del of untranslocated TEL, 3—single fusion loss of AML1/TEL fusion on der 12p, 4—extra copies of AML 1(number), 5—extra copies of TEL (number), additional genetic abnormality: +, 6—clones (Number), clone size (positivity rate). Normal/abnormal karyotype: NK/AK, inadequate/no metaphases: IM/NM, *Ploidy analysis—chromosome number.
Case16: 46,XY,dup(17)(q21q22),t(?;17)(?;q25)[3]/47,XY, + 11,dup(17)(q21q22)t(?;17)(?;q25)[4]/46,XY,del(11)(q23),dup(17)(q21q22)t(?;17)(?;q25)[3]/46,XY,dup(17)(q21q22)t(?;17)(?;q25),t(?;18)(?;p11.2)[3]/46,XY[7].
Case18: 46,XY,inv(11)(p13q13.1)[1]/46,XY,inv(11)(p13q13.1),del(17)(p11.2)[5]/46,XY,inv(11)(p13q13.1),del(12)(p13)[4]/ 46,XY,inv(11)(p13q13.1),del(12)(p13),del(17)(p11.2)[8]/46,XY[2].
|
Parameters |
Group 1: 1–18 years |
Group 2: 19–39 years |
Group 3: >40 years |
|
|---|---|---|---|---|
|
Overall, no of patients (%) |
125(58.41) |
63(29.43) |
26(12.14) |
|
|
Median age of presentation (y) |
5 |
26 |
46.5 |
|
|
No of males No of females Gender of baby not disclosed |
77 47 1 |
45 18 |
15 11 |
|
|
Male female ratio |
1.6:1 |
2.5:1.0 |
1.3:1 |
|
|
TEL AML1 fusion positive cases |
Total:18/214 (8.41%), FISH:15/178(8.6%) / RT-PCR:3/36(8.3%) |
|||
|
TEL AML1 fusion positive cases as per age group by both technologies: No (%) |
17/125(13.6%) |
1/63(1.58%) |
– |
|
|
TEL AML1 negative cases:196/214(91.58%) |
TELAML1negative with additional genetic findings:39/163(23.9%) |
|||
|
Types of additional abnormalities in negative group No of patients (%) &%-cells with abnormal findings in different patients. |
||||
|
Additional copies of AML1—21(12.8%) 24–92 | ||||
|
Case no |
Cytogenetic findings |
FISH findings |
Correlation of karyotype and FISH findings |
|---|---|---|---|
|
1 |
47,XX,+8[14],46X-X-[2]* |
TEL/AML1 negative, with amplification of the AML1 gene—2 copies in 45%-cells |
Cryptic amplification of AML1 gene |
|
2 |
50–57,XX/XXX,+X[11],+4[10],+5[6],+6[13],+8[6],+9[7],+10[6],+11[8],+12[3],t(?;12)(?;q24.1)[5],+14[4],+15[2],+16[3],+17[12],+18[6],−9[11],+20[3],+21[10][cp16]/46,XX[4] |
TEL/AML1 negative, amplification of AML1 gene (1 copy) |
Karyotype demonstrating a+21 corroborating with FISH amplification. |
|
3 |
46,XY,dup(9)(q34.1q34.1)[4]/47,XY,+mar[2]/46,XY[9] |
TEL/AML1 negative, amplification of AML1 gene1–2 copies -33%- |
Cryptic amplification of AML1 seen. |
|
4 |
46,XY,del(8)(p22)[8]/46,XY[6] |
TEL/AML1 negative, Amplification of Tel and AML1 genes |
Cryptic amplification of TEL and AML1 genes |
|
5 |
50–53,XXY/XY,+X,+4,+6,+8,+10,+14,+17,+18, +21,+22[cp15]/46,XY[5] |
TEL/AML1 negative, amplification of AML1 gene—2 copies |
Karyotype shows gain of chromosome 21 (only 1 copy) and the other copy of AML 1 is cryptic. |
|
6 |
54–55,XY,+4[4],+6[2],+8[3],+10[3],+16[1],+17 [3],+17×2[1],+18[3],+21[2],+21×2[2],+22[3][cp4]/46,XY[16]13 |
TEL/AML1 negative, amplification of AML1 gene—2 copies—58%- |
Karyotype and FISH demonstrate gain of 2 copies of chromosome 21 and 2 copies of AML1 gene |
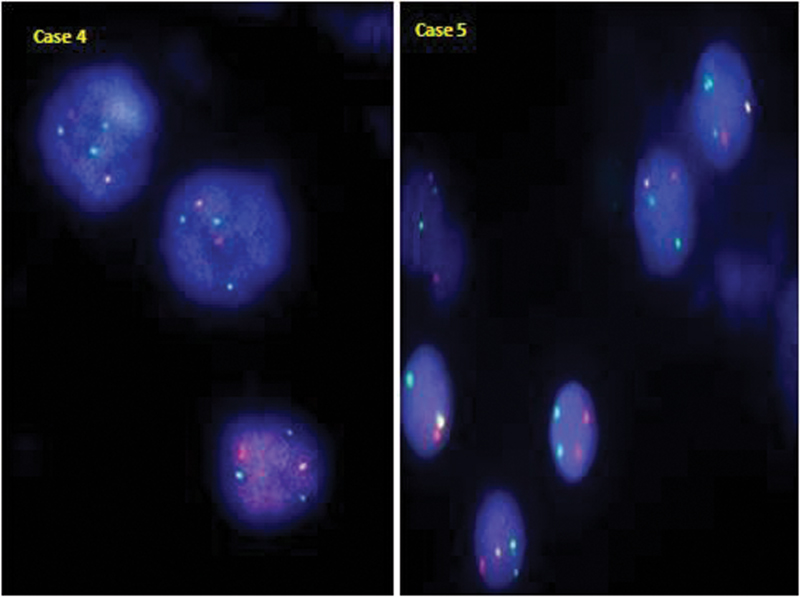
| Fig 1 :TEL/AML1 positive case with an atypical signal pattern showing 1 fusion with additional copies of AML1 (green) and TEL (orange) gene in case 4 and an additional copy of only AML1 gene (green) in case 5 (clone 2).
TEL/AML1 fusion was absent in 91.58%-cases. Additional genetic abnormalities were also noted in 23.9%-of these patients, with amplification of AML1, seen more often ([Table 2]).
Cytogenetic analysis in 92 B-ALL patients revealed the karyotype was normal in 57 (61.95%) and abnormal in 35 /92 (38.04%) patients. It is important to note that 14 patients with normal karyotype and cases 16 and 18 with abnormal complex cytogenetics had TEL/AML1 rearrangement/ mutation. Case 18 showed evidence of del(12)(p13)in 60%-metaphases ([Table 1]-footnote).
Correlation of FISH results and cytogenetics demonstrated TEL/AML1 rearrangement to be the sole abnormality and mutually exclusive as BCR ABL and MLL were negative for rearrangement in all cases wherever done. Additional genetic abnormalities noted in the positive group, occurred when the chromosome complement was 46 with two intact homologous chromosomes 12 and 21 suggesting that the abnormalities were submicroscopic/cryptic level.
Among the 39 cases negative for TEL/AML1 rearrangement but with additional abnormalities, karyotype was normal in 8 and abnormal in 6 cases. Correlation studies demonstrated that the loss or gain of the genes was cryptic in the former. However, in six cases having an abnormal karyotype, AML1 amplification was cryptic in 50%-and corroborated by additional chromosome 21 ([Table 3]).
Complete blood count (CBC) in patients positive for TEL/AML1fusion revealed low hemoglobin in all patients (range: 35–116 g/L, median: 74 g/L), high white blood cell (WBC) counts in 8/18 (44.4%) and 7/17(41.2%) pediatric patients (range: 11.2–110 × 109/L) and low counts in 4/18(22.2%) and 4/17 pediatric patients (23.52%) (range: 1.01–3.6). Platelet counts were low in all patients (range: 10–115 × 109/L, median: 42 × 109/L). The blasts ranged from 0 to 98% (median: 41%) in peripheral smear and 0 to 95% (median: 80%) in the bone marrow ([Fig. 2]). Immunophenotyping revealed patients had CD34 positive phenotype, common acute lymphoblstic leukemia associated antigen (CALLA) positive in 17/18 (94.4%) and aberrant expression of CD13, CD33, CD56, singly or in combination in 10/17(58.8%) patients. Coexpression of CD13 and CD33 occurred in 70% ([Fig. 3]), only CD3 in 20%, and only CD56 in 10%.
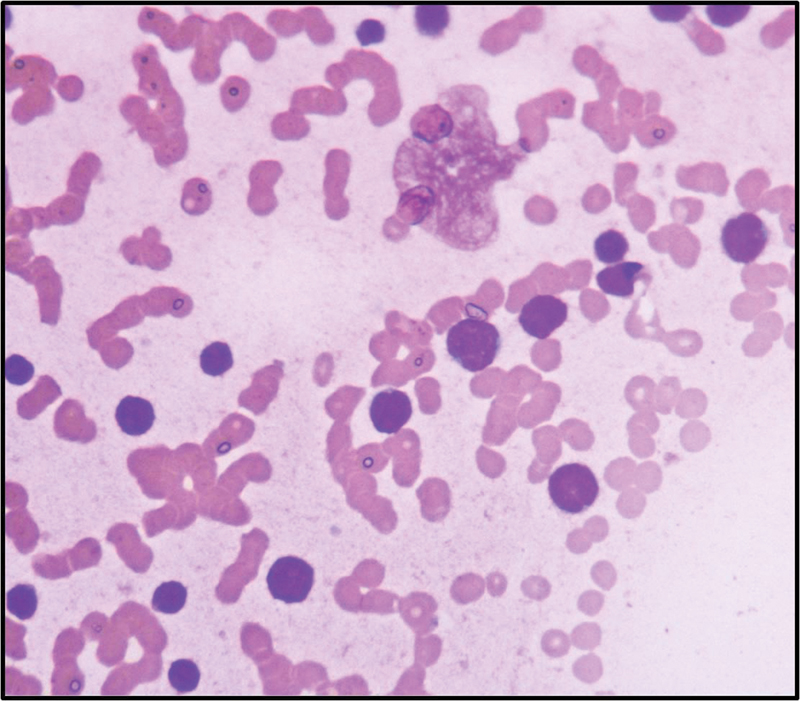
| Figure 2: Blasts showing high N/C ratio coarse chromatin and scanty cytoplasm (Leishman stain,400x).
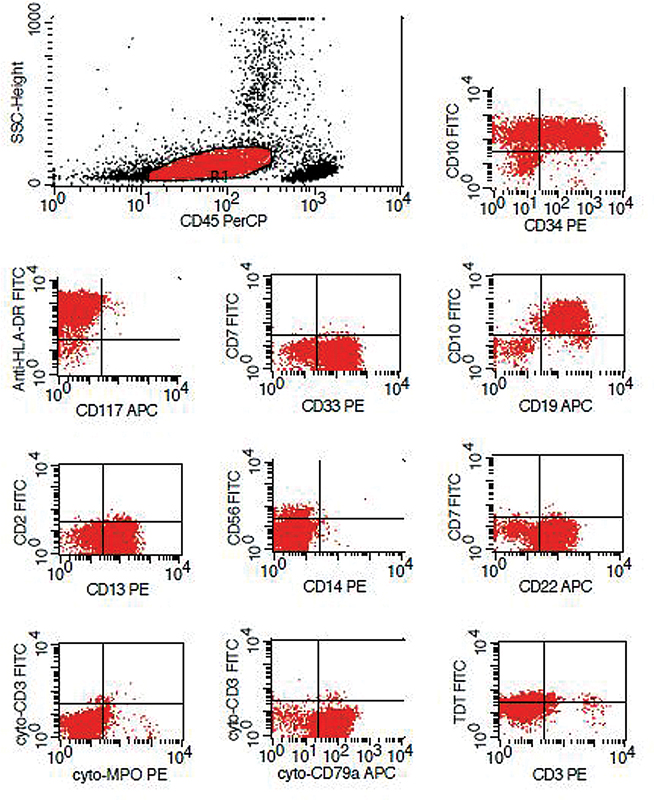
| Figure 3: Immunophenotyping in a prototype case showing aberrant expression of CD13 and CD33.
Treatment and follow-up details: All the 18 patients were treated with ALL-Berlin-Frankfurt-Munster (BFM 95) protocol. One child died after 1 month due to thrombotic complication. Three patients completed induction and consolidation and lost to follow-up. Fourteen patients who completed the whole treatment are alive and disease free. The median survival among those who completed treatment is 35.5 months (range: 19–44 months).
Discussion
ALL is the most frequent malignancy accounting for 85%-of all childhood leukaemia.[32] [33] [34] [35] [36] B-ALL constituted 76%-of all ALL that comprised of predominantly CALLA positive ALL in 73%, early precursor pro B-ALL in 3%.[33] The World Health Organization classification of hematopoietic and lymphoid tissue (2017) defines B lymphoblastic leukemia/lymphoma into various subtypes including “B lymphoblastic leukemia/lymphoma with 7 recurrent genetic abnormalities.”[1] Conventional cytogenetics, interphase FISH, RT-PCR find application in the detection of recurrent genetic abnormalities as they play a pivotal role in prognostication, selection of therapy, and response to treatment.[1] [13] [31] [32] [33] [34] [35] [36] [37]
Translocation t(12;21)(p13;q22), a somatically acquired early lesion, results in the critical TEL/AML1 fusion on chromosome 21, and AML1/ TEL fusion on derivative 12p that can be lost. TEL allele is often deleted the fusion protein acts in a dominant negative fashion inhibiting the normal function of transcription factor AML1.[4] The abnormality can occur as a sole abnormality or within the context of a complex karyotype characterized by three or more chromosomal abnormalities. Conventional cytogenetics is the gold standard to detect both known and unknown genomic abnormalities in the genome; however, poor chromosome morphology, few malignant metaphases encountered in ALL, and inherent cytogenetic invisibility of the abnormality limit the detection of t(12;21) to 0.05%-of patients. Cryptic and subtle abnormalities, below 7 to 8 MB resolution, can be missed on karyotyping. FISH and RT-PCR enable detection of TEL/AML1 in 20 to 25%-of precursor B ALL.[2] Translocation t(12;21) has a favorable outcome and useful for the prediction of outcome in childhood ALL. Coexistence of the same, with other gene rearrangements and in complex setting, favors poorer clinical outcome.
B-ALL patients with t(12;2l) abnormality are younger, ∼3–5 years, blasts have B precursor phenotype, and are CD19, CD10 and CD34 positive with expression of myeloid antigens CD13, sensitive to chemotherapy and associated with favorable prognosis and late relapse.[1] [2] [3] [33] TEL/AML1 fusion is seen rarely in adults (1–4%);[2] [3] however, increased numbers of adults diagnosed with B ALL, prompted assessment for the same. In the present study, under-representation of the cryptic TEL/AML1 rearrangement overall (8.4%) and in children (13.6%) is low contrary to the expected high prevalence of 25%-in western countries.[4] Hospital-based reports,[5] [6] [7] [9] [12] [13] [14] [15] [16] [17] [18] [19] [20] [21] [22] [23] [24] [25] [26] [27] [28] [29] [30] international projects,[1] [10] [24] multicenter studies,[11] [24] and review articles[8] [15] indicated existence of geographical differences in distribution of TEL/AML1 worldwide, including India ([Table S1] and [S2]). Global studies indicated the incidence of TEL/AML1 positive B ALL was high ranging from 17%-in Spain,[6]18.9%-in Brazil,[17]28%-in Tunisia,[7] 31%-in United States,[5] 37.5%-in Egypt,[14] to 40%-in Israel.[18] Exception was Turkey (8.5%).[16] The incidence was low in Asian countries, 14.1 to 17.9%-in China and Korea,[13] [14] [15] 5 to 10.6% in Pakistan,[10] [11] [12] and 0 to 19.8%-in north and western India,[19] [20] [21] [22] [23] [24] [25] [26] 8.3%-in Chennai,[27]4.3%-in Karnataka,[28]4.8%-in Kerala,[29] and 13.64%-in Hyderabad.[30] The low prevalence of TEL/AML1 positivity is 13.6%-children with B-ALL, which resonates with previous studies from this region.[27] [30] Variation in the incidence of TEL/AML1 positivity may be dependent on the technology used. When conventional cytogenetics and FISH were used for testing, the global incidence varied from 8.3 to 19.8%[13] [17] [25] [26] [27] however, when only FISH was used it varied from 8.5 to 40%,[5] [6] [7] [9] [14] [16] [18] [28] [30] and 4.3 to 13.64%-in south India.[28] [30] RT-PCR enabled detection of patients with the abnormality in 7.4 to 10.8%-in developing countries like Pakistan[10] [11] [12] and in 0 to 16.2%-in India.[19] [20] [21] [22] [24] [29] Preselection of precursor B ALL enabled increased detection of the TEL/AML1 rearranged cases. Affordability for the test is an important factor. In the present study, both technologies enabled detection of the fusion gene at the same rate of 8.3 to 8.4%. The population studied, sample size, patient selection, technology(s) used for testing affect the detection rate. If FISH is the modality, the probes used in house/commercial, company, design of the probe, size, validation, cutoff value set, and interpretation are important for defining the status of the abnormality. For RT-PCR, the primers were used.
It is well known that reciprocal translocation t(12;21)(p13;q22) results in the pathogenic TEL/AML1 fusion on chromosome 21, and AML1/TEL fusion on derivative 12p.The latter may be lost sometimes. Deletion of TEL allele on the normal chromosome as a consequence of del(12)(p12) and duplication of der(21)t(12;21) that looks like a +21 hyperdiploid karyotype with <5 class="i" xss=removed>AML1 and rarely gene amplification with cluster of >5 or intra-chromosomal amplification of AML1 is also encountered. Interphase FISH enabled the detection of not only TEL/AML1 fusion but also associated additional genetic abnormalities in 93%-of these cases and in the negative group too. The reported loss of the untranslocated TEL,[5] [7] [14] [16] [25] [27] duplication of TEL/AML1 fusion (der 21t(12;21)) were seen infrequently in our study as compared with 35 and 17%, respectively, reported previously.[25] An unusual finding was the occurrence of cryptic loss of the reciprocal AML1-TEL fusion on der12p in 66.5%-of our patients. This appears to be rare[8] and has not been reported earlier from this region. Loss of derivative 12p was also associated with extra copy(s) of AML1 in 40%, with amplification of TEL gene (33.3%). These findings were consistent with the previous reports.[5] [6] [7] [8] [13] [14] [16] [17] [18] TEL/AML1 rearrangement with one or more copies of der 21t(12;21) has been reported in 42 to 50%-with additional abnormalities,[5] [6] [8] [13] [17] [18] [38] including additional AML1 in 6 to 23.8%-patients,[5] [6] [7] [14] [16] [25] and clones with loss of TEL gene and an extra AML1 in 9.5%.[6] Duplication of the fusion gene is rare and may occur in the form of duplication of der 21t(12;21) or ider 21(q10)t(12;21). TEL gene functions as a tumor suppressor gene, and its function may be inhibited when in combination with TEL/AML1 fusion in a dominant negative fashion. Alternatively, TEL gene may inactivate the fusion protein necessitating the deletion of the TEL gene for leukemogenesis. Submicroscopic deletions and point mutations of TEL homologue may contribute to inactivation of TEL.[25] Additional genetic abnormalities in relation to the cryptic t(12;21) translocation are envisaged to be secondary events critical to leukemogenesis and not disease progression. The significance of additional genetic changes with respect to clinical features and outcome between presence or absence is debatable.[14]
The observation of TEL/AML1 rearrangement and additional genetic abnormalities detected by FISH in patients having normal cytogenetics in 14/18 (77%) patients, complex karyotype in 2 (11.11%), one with 12p deletion in 80%-metaphases connoting loss of AML1/TEL fusion, and the other case with nonrandom chromosomal abnormalities indicate (1) TEL/AML1 rearrangement is the sole, cryptic abnormality; (2) additional mutational events were submicroscopic too, and (3) the normal karyotype is actually a pseudodiploid karyotype.
Evaluation of clinical and pathological features demonstrated occurrence of TEL/AML1 positivity mainly in children (94.4%), aged 2 to 13 years (median: 4 years) predominantly among males (94.1%). The fusion was absent in all adults tested. The patients had classic CD34 and CD10 positive phenotype (CALLA positive B-ALL) with aberrant expression of cross lineage antigens, CD13 and CD33.[2] [33] WBC counts were high in 44.4%-patients, low in 22.2%, and rest within normal range. Another study demonstrated significant association of TEL/AML1 positive group with lower age group > 1 to 5 years (median: 5 years), WBC counts of 10–20 × 109/L (median: WBC count of 11.2 ×109/L), CD34 negative phenotype, and absence of CD13/33 myeloid marker expression and low relapse rate.[25]
Our hospital is a regional tertiary care cancer center for cancer treatment in a metropolitan city. The possibility of geographic variation in presentation of the disease cannot be ruled out. India is genetically and ethnically a diverse country. Genetic propensity for TEL/AML1 fusion in childhood ALL has been described.[37] Regional, population based, biological differences, gene–environment interactions, are critical in leukemogenesis and may differentially contribute to the prevalence of TEL/AML1 fusion occurrence.
This study revealed additional genetic abnormalities in 23.9%-TEL/AML1 negative patients lacking the BCR-ABL and MLL gene rearrangement. These include extra AML1 copies, extra AML1, alone or in combination with TEL aberrations (amplification or deletion) and abnormalities of TEL gene ([Table 2]). Extra AML1 was cryptic or corroborated by cytogenetics ([Table 2]). One study reported no loss of TEL homologue, extra copies of AML1 in 37%. The authors cited that AML1 may be the current target of genetic abnormalities in ALL and acute myeloid leukemia, which probably induces the transcriptional defect in AML1 in the process of leukemogenesis, and showed abnormalities. These occur as a consequence of “tri/tetra/polysomy 21, high degree of amplification, and jumping translocations/insertions.” The biological significance of the observed anomalies is not known. Although we have not analyzed these cases for clincopathological correlations, TEL/AML1 negative cases with extra AML1/trisomy 21 were reported to be associated with CD13/33-negative and CD34-positive phenotype, low LDH value, low WBC counts, age ranging from 1 to 5 years, high hyperploidy.[25] B-ALL with iAMP21[1] has been defined as the presence of 3 or more RUNX1 signals on one marker chromosome or a total of five or more RUNX1 signals per cell. Metaphase FISH with Vysis TEL/AML1 dual color dual fusion probes display one signal connoting a normal chromosome 21, and several red signals along an abnormal chromosome 21 as tandem duplication, while interphase FISH displays a cluster of red signals (AML1) and one separate red signal representing the normal chromosome 21.[1] [38] [39] To the best of our knowledge, interphase FISH showed distinct well separated signals of AML1, and no localized clusters in all with multiple copies of AML1. However as per the definition, three patients with five or more AML1 signals per cell can be construed as having an iAMP amplification. It is important to identify patients with iAMP who have a poor prognosis.
Each technology has its advantages and limitations. Prevalence of good prognostic TEL/AML1 positive patients was found to be low at this center. Multicenter studies should be undertaken to understand this genetic subtype of B-ALL better in larger sample volume and prognostic implications.
The recurrent genetic abnormalities tested in B-ALL patients have revealed large number of patients lacking these abnormalities and need further interrogation. Comprehensive emerging technologies have identified several genetic determinants. Next-generation sequencing and other evolving technologies will address these genetic abnormalities. Hence, use of orthogonal testing and algorithmic approach will propel identification of new entities in our population.
Conclusion
This is the first hospital-based large study from south India demonstrating low prevalence of cryptic TEL/AML1 in 13.6%-children with BALL, lower than western data but comparable with south Indian studies. FISH was useful in identifying unique additional genetic abnormalities in TEL/AML1 positive and negative cases. Clinicopathologic associations were classic. The study adds to the Indian data. The detection of relevant specific abnormalities in B-ALL patients is of fundamental importance.
Conflict of Interest
None declared.
Acknowledgments
Authors would like to thank the contribution of the DNB residents who performed bone marrow aspiration and biopsy for evaluation and diagnosis, Mr. Janaiah, and Mr. K Ramchander Reddy for technical support for enabling CBC, flowcytometry, conventional cytogenetics, and Mrs. M Padma Rani for processing blood/bone marrow samples for FISH analysis.
Supplementary MaterialReferences
References

| Fig 1 :TEL/AML1 positive case with an atypical signal pattern showing 1 fusion with additional copies of AML1 (green) and TEL (orange) gene in case 4 and an additional copy of only AML1 gene (green) in case 5 (clone 2).

| Figure 2: Blasts showing high N/C ratio coarse chromatin and scanty cytoplasm (Leishman stain,400x).

| Figure 3: Immunophenotyping in a prototype case showing aberrant expression of CD13 and CD33.


 PDF
PDF  Views
Views  Share
Share

